Mol:FL1C3CGS0011
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 44 0 0 0 0 0 0 0 0999 V2000 | + | 41 44 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.5503 -1.8803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5503 -1.8803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5503 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5503 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8354 -3.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8354 -3.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1205 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1205 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1205 -1.8803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1205 -1.8803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8354 -1.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8354 -1.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5939 -3.1183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5939 -3.1183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3069 -2.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3069 -2.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0183 -3.1174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0183 -3.1174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7282 -2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7282 -2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4230 -3.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4230 -3.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1175 -2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1175 -2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1175 -1.9054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1175 -1.9054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4230 -1.5043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4230 -1.5043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7282 -1.9054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7282 -1.9054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5939 -3.9416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5939 -3.9416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8354 -3.9437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8354 -3.9437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2648 -1.4678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2648 -1.4678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8122 -1.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8122 -1.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1761 -3.1839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1761 -3.1839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8122 -3.1086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8122 -3.1086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7708 -0.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7708 -0.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9535 -0.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9535 -0.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3886 0.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3886 0.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3759 0.9209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3759 0.9209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1932 1.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1932 1.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7581 0.4143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7581 0.4143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0888 2.0271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0888 2.0271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5446 1.7045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5446 1.7045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6537 -0.2207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6537 -0.2207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5418 3.3094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5418 3.3094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3826 3.3094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3826 3.3094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8122 2.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8122 2.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9724 2.3622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9724 2.3622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0896 2.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0896 2.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3826 2.9311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3826 2.9311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5418 3.7781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5418 3.7781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0973 3.9437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0973 3.9437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8122 3.5309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8122 3.5309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5292 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5292 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1952 1.9545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1952 1.9545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 3 17 1 0 0 0 0 | + | 3 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 20 2 1 0 0 0 0 | + | 20 2 1 0 0 0 0 |
| − | 12 21 1 0 0 0 0 | + | 12 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 23 18 1 0 0 0 0 | + | 23 18 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 35 28 1 0 0 0 0 | + | 35 28 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 27 40 1 0 0 0 0 | + | 27 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 42 | + | M SBL 1 1 42 |
| − | M SMT 1 HOH2^C | + | M SMT 1 HOH2^C |
| − | M SBV 1 42 0.0000 -0.6736 | + | M SBV 1 42 0.0000 -0.6736 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 44 0.7711 -0.2939 | + | M SBV 2 44 0.7711 -0.2939 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL1C3CGS0011 | + | ID FL1C3CGS0011 |
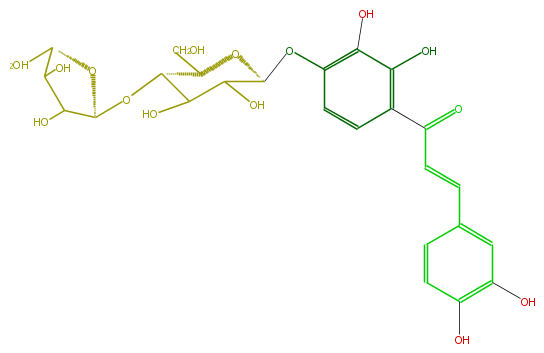
| − | FORMULA C26H30O15 | + | FORMULA C26H30O15 |
| − | EXACTMASS 582.15847029 | + | EXACTMASS 582.15847029 |
| − | AVERAGEMASS 582.5074 | + | AVERAGEMASS 582.5074 |
| − | SMILES c(O)(c2OC(C(O)4)OC(C(C(O)4)OC(O3)C(C(C3CO)O)O)CO)c(c(cc2)C(C=Cc(c1)ccc(c1O)O)=O)O | + | SMILES c(O)(c2OC(C(O)4)OC(C(C(O)4)OC(O3)C(C(C3CO)O)O)CO)c(c(cc2)C(C=Cc(c1)ccc(c1O)O)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 44 0 0 0 0 0 0 0 0999 V2000
-1.5503 -1.8803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5503 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8354 -3.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1205 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1205 -1.8803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8354 -1.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5939 -3.1183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3069 -2.7067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0183 -3.1174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7282 -2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4230 -3.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1175 -2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1175 -1.9054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4230 -1.5043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7282 -1.9054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5939 -3.9416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8354 -3.9437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2648 -1.4678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8122 -1.5045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1761 -3.1839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8122 -3.1086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7708 -0.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9535 -0.6557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3886 0.0700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3759 0.9209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1932 1.1399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7581 0.4143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0888 2.0271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5446 1.7045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6537 -0.2207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5418 3.3094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3826 3.3094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8122 2.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9724 2.3622 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0896 2.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3826 2.9311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5418 3.7781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0973 3.9437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8122 3.5309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5292 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1952 1.9545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
3 17 1 0 0 0 0
1 18 1 0 0 0 0
13 19 1 0 0 0 0
20 2 1 0 0 0 0
12 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
23 18 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 31 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
35 28 1 0 0 0 0
38 39 1 0 0 0 0
33 39 1 0 0 0 0
40 41 1 0 0 0 0
27 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 42
M SMT 1 HOH2^C
M SBV 1 42 0.0000 -0.6736
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 44
M SMT 2 ^ CH2OH
M SBV 2 44 0.7711 -0.2939
S SKP 5
ID FL1C3CGS0011
FORMULA C26H30O15
EXACTMASS 582.15847029
AVERAGEMASS 582.5074
SMILES c(O)(c2OC(C(O)4)OC(C(C(O)4)OC(O3)C(C(C3CO)O)O)CO)c(c(cc2)C(C=Cc(c1)ccc(c1O)O)=O)O
M END