Mol:FL1CDAGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 33 35 0 0 0 0 0 0 0 0999 V2000 | + | 33 35 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.7155 0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7155 0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7155 -0.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7155 -0.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1615 -0.5669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1615 -0.5669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6075 -0.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6075 -0.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6075 0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6075 0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1615 0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1615 0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0538 -0.5668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0538 -0.5668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5013 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5013 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9500 -0.5661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9500 -0.5661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3998 -0.2484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3998 -0.2484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1386 -0.5593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1386 -0.5593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6769 -0.2484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6769 -0.2484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6769 0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6769 0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1386 0.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1386 0.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3998 0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3998 0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0538 -1.2048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0538 -1.2048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0711 0.6258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0711 0.6258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1615 -1.2063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1615 -1.2063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1457 0.9590 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.1457 0.9590 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.8449 0.4380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.8449 0.4380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4234 0.6033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4234 0.6033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0053 0.4380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.0053 0.4380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.3062 0.9590 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.3062 0.9590 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7277 0.7937 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.7277 0.7937 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.5870 0.8093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5870 0.8093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7692 1.2683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7692 1.2683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8894 0.8028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8894 0.8028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0538 0.7123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0538 0.7123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3393 0.2998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3393 0.2998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0728 1.0114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0728 1.0114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5728 1.8774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5728 1.8774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2049 0.1456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2049 0.1456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2049 -0.6794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2049 -0.6794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 17 13 1 0 0 0 0 | + | 17 13 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 5 28 1 0 0 0 0 | + | 5 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 22 32 1 0 0 0 0 | + | 22 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 32 33 | + | M SAL 3 2 32 33 |
| − | M SBL 3 1 34 | + | M SBL 3 1 34 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 34 3.3583 0.4647 | + | M SVB 3 34 3.3583 0.4647 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 30 31 | + | M SAL 2 2 30 31 |
| − | M SBL 2 1 32 | + | M SBL 2 1 32 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 32 -4.0728 1.0114 | + | M SVB 2 32 -4.0728 1.0114 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 28 29 | + | M SAL 1 2 28 29 |
| − | M SBL 1 1 30 | + | M SBL 1 1 30 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 30 -2.0538 0.7123 | + | M SVB 1 30 -2.0538 0.7123 |
| − | S SKP 8 | + | S SKP 8 |
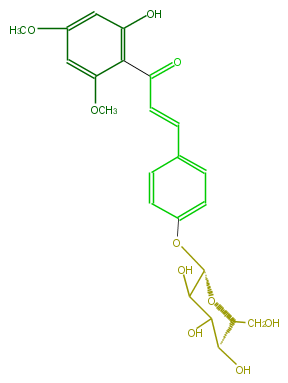
| − | ID FL1CDAGS0001 | + | ID FL1CDAGS0001 |
| − | KNApSAcK_ID C00007888 | + | KNApSAcK_ID C00007888 |
| − | NAME 4,2'-Dihydroxy-4',6'-dimethoxychalcone 4-glucoside | + | NAME 4,2'-Dihydroxy-4',6'-dimethoxychalcone 4-glucoside |
| − | CAS_RN 68984-71-4 | + | CAS_RN 68984-71-4 |
| − | FORMULA C23H26O10 | + | FORMULA C23H26O10 |
| − | EXACTMASS 462.152597052 | + | EXACTMASS 462.152597052 |
| − | AVERAGEMASS 462.44654 | + | AVERAGEMASS 462.44654 |
| − | SMILES c(c3)(c(c(cc3OC)O)C(=O)C=Cc(c2)ccc(c2)O[C@@H](C(O)1)O[C@H](CO)[C@H](O)C1O)OC | + | SMILES c(c3)(c(c(cc3OC)O)C(=O)C=Cc(c2)ccc(c2)O[C@@H](C(O)1)O[C@H](CO)[C@H](O)C1O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
33 35 0 0 0 0 0 0 0 0999 V2000
-3.7155 0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7155 -0.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1615 -0.5669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6075 -0.2471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6075 0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1615 0.7125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0538 -0.5668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5013 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9500 -0.5661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3998 -0.2484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1386 -0.5593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6769 -0.2484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6769 0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1386 0.6840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3998 0.3732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0538 -1.2048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0711 0.6258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1615 -1.2063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1457 0.9590 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.8449 0.4380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4234 0.6033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0053 0.4380 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.3062 0.9590 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7277 0.7937 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.5870 0.8093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7692 1.2683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8894 0.8028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0538 0.7123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3393 0.2998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0728 1.0114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5728 1.8774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2049 0.1456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2049 -0.6794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
17 13 1 0 0 0 0
3 18 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
17 20 1 0 0 0 0
5 28 1 0 0 0 0
28 29 1 0 0 0 0
1 30 1 0 0 0 0
30 31 1 0 0 0 0
22 32 1 0 0 0 0
32 33 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 32 33
M SBL 3 1 34
M SMT 3 CH2OH
M SVB 3 34 3.3583 0.4647
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 30 31
M SBL 2 1 32
M SMT 2 OCH3
M SVB 2 32 -4.0728 1.0114
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 28 29
M SBL 1 1 30
M SMT 1 OCH3
M SVB 1 30 -2.0538 0.7123
S SKP 8
ID FL1CDAGS0001
KNApSAcK_ID C00007888
NAME 4,2'-Dihydroxy-4',6'-dimethoxychalcone 4-glucoside
CAS_RN 68984-71-4
FORMULA C23H26O10
EXACTMASS 462.152597052
AVERAGEMASS 462.44654
SMILES c(c3)(c(c(cc3OC)O)C(=O)C=Cc(c2)ccc(c2)O[C@@H](C(O)1)O[C@H](CO)[C@H](O)C1O)OC
M END