Mol:FL2FAAGS0014
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6761 0.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6761 0.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6761 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6761 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1198 -0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1198 -0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4365 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4365 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4365 0.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4365 0.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1198 1.0431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1198 1.0431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9928 -0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9928 -0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5491 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5491 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5491 0.7219 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.5491 0.7219 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9928 1.0431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9928 1.0431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1052 1.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1052 1.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6721 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6721 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2391 1.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2391 1.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2391 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2391 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6721 2.0250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6721 2.0250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1052 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1052 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9928 -0.7854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9928 -0.7854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2324 1.0431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2324 1.0431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8061 2.0250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8061 2.0250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1198 -0.8838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1198 -0.8838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2964 0.9097 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.2964 0.9097 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.9283 0.4238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.9283 0.4238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3981 0.6299 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.3981 0.6299 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8866 0.6354 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8866 0.6354 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2583 1.0072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2583 1.0072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7221 0.7625 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.7221 0.7625 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.8059 1.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8059 1.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4365 0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4365 0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0944 0.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0944 0.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1002 -1.2353 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.1002 -1.2353 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.7320 -1.7212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.7320 -1.7212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.2019 -1.5151 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.2019 -1.5151 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.6903 -1.5096 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.6903 -1.5096 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.0621 -1.1378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0621 -1.1378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5259 -1.3825 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.5259 -1.3825 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -2.6862 -1.1840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6862 -1.1840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0795 -2.1788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0795 -2.1788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8981 -2.0250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8981 -2.0250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9234 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9234 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8750 2.0090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8750 2.0090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7272 -0.4434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7272 -0.4434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6788 -0.1360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6788 -0.1360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 26 39 1 0 0 0 0 | + | 26 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 45 -1.7272 -0.4434 | + | M SVB 2 45 -1.7272 -0.4434 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 43 -2.9234 1.7016 | + | M SVB 1 43 -2.9234 1.7016 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FAAGS0014 | + | ID FL2FAAGS0014 |
| − | KNApSAcK_ID C00008214 | + | KNApSAcK_ID C00008214 |
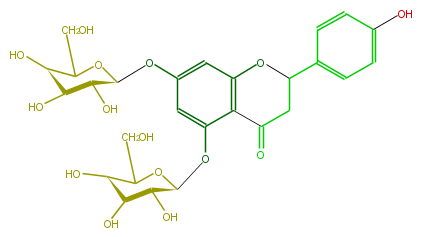
| − | NAME Naringenin 5,7-di-O-glucoside | + | NAME Naringenin 5,7-di-O-glucoside |
| − | CAS_RN 5180-65-4 | + | CAS_RN 5180-65-4 |
| − | FORMULA C27H32O15 | + | FORMULA C27H32O15 |
| − | EXACTMASS 596.174120354 | + | EXACTMASS 596.174120354 |
| − | AVERAGEMASS 596.5339799999999 | + | AVERAGEMASS 596.5339799999999 |
| − | SMILES C(C(O1)[C@@H]([C@H](O)[C@H](O)[C@H](Oc(c4)cc(O2)c(c4O[C@H](O5)[C@H]([C@@H](O)[C@H](C5CO)O)O)C(=O)CC2c(c3)ccc(c3)O)1)O)O | + | SMILES C(C(O1)[C@@H]([C@H](O)[C@H](O)[C@H](Oc(c4)cc(O2)c(c4O[C@H](O5)[C@H]([C@@H](O)[C@H](C5CO)O)O)C(=O)CC2c(c3)ccc(c3)O)1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.6761 0.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6761 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1198 -0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4365 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4365 0.7219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1198 1.0431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9928 -0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5491 0.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5491 0.7219 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9928 1.0431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1052 1.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6721 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2391 1.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2391 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6721 2.0250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1052 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9928 -0.7854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2324 1.0431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8061 2.0250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1198 -0.8838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2964 0.9097 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.9283 0.4238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3981 0.6299 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8866 0.6354 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2583 1.0072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7221 0.7625 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.8059 1.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4365 0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0944 0.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1002 -1.2353 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.7320 -1.7212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.2019 -1.5151 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6903 -1.5096 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.0621 -1.1378 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5259 -1.3825 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-2.6862 -1.1840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0795 -2.1788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8981 -2.0250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9234 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8750 2.0090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7272 -0.4434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6788 -0.1360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
18 1 1 0 0 0 0
14 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 20 1 0 0 0 0
26 39 1 0 0 0 0
39 40 1 0 0 0 0
35 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 CH2OH
M SVB 2 45 -1.7272 -0.4434
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 CH2OH
M SVB 1 43 -2.9234 1.7016
S SKP 8
ID FL2FAAGS0014
KNApSAcK_ID C00008214
NAME Naringenin 5,7-di-O-glucoside
CAS_RN 5180-65-4
FORMULA C27H32O15
EXACTMASS 596.174120354
AVERAGEMASS 596.5339799999999
SMILES C(C(O1)[C@@H]([C@H](O)[C@H](O)[C@H](Oc(c4)cc(O2)c(c4O[C@H](O5)[C@H]([C@@H](O)[C@H](C5CO)O)O)C(=O)CC2c(c3)ccc(c3)O)1)O)O
M END