Mol:FL3FAAGS0034
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.2539 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2539 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2539 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2539 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8102 -0.5363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8102 -0.5363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3665 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3665 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3665 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3665 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8102 0.7484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8102 0.7484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9228 -0.5363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9228 -0.5363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4791 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4791 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4791 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4791 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9228 0.7484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9228 0.7484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9228 -1.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9228 -1.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0352 0.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0352 0.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6022 0.4209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6022 0.4209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1692 0.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1692 0.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1692 1.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1692 1.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6022 1.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6022 1.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0352 1.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0352 1.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3022 0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3022 0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8102 -1.1784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8102 -1.1784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7360 1.7302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7360 1.7302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6147 0.6161 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.6147 0.6161 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.1801 0.0424 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1801 0.0424 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5541 0.2858 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.5541 0.2858 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.9502 0.2923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.9502 0.2923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.3890 0.7313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3890 0.7313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0284 0.5017 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.0284 0.5017 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.5031 0.2828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5031 0.2828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8577 0.0094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8577 0.0094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1955 -0.3163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1955 -0.3163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1608 0.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1608 0.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7360 0.2622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7360 0.2622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7360 -0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7360 -0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1608 -0.7340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1608 -0.7340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5856 -0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5856 -0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1608 1.0888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1608 1.0888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1608 -1.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1608 -1.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5856 -1.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5856 -1.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0104 -1.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0104 -1.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0104 -0.7340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0104 -0.7340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4353 -1.7303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4353 -1.7303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2689 1.2346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2689 1.2346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6019 1.9797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6019 1.9797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 30 35 2 0 0 0 0 | + | 30 35 2 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 45 -2.2689 1.2346 | + | M SVB 1 45 -2.2689 1.2346 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0034 | + | ID FL3FAAGS0034 |
| − | KNApSAcK_ID C00004170 | + | KNApSAcK_ID C00004170 |
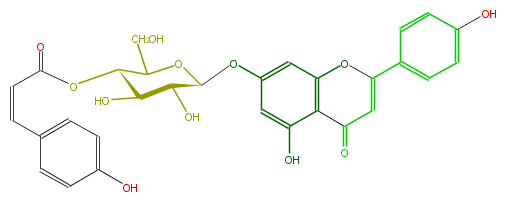
| − | NAME Apigenin 7-(4''-Z-p-coumarylglucoside) | + | NAME Apigenin 7-(4''-Z-p-coumarylglucoside) |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C30H26O12 | + | FORMULA C30H26O12 |
| − | EXACTMASS 578.1424262959999 | + | EXACTMASS 578.1424262959999 |
| − | AVERAGEMASS 578.5202400000001 | + | AVERAGEMASS 578.5202400000001 |
| − | SMILES O[C@H]([C@@H]1O)[C@@H](OC(C=Cc(c5)ccc(c5)O)=O)C(O[C@H]1Oc(c4)cc(c(c43)C(=O)C=C(O3)c(c2)ccc(O)c2)O)CO | + | SMILES O[C@H]([C@@H]1O)[C@@H](OC(C=Cc(c5)ccc(c5)O)=O)C(O[C@H]1Oc(c4)cc(c(c43)C(=O)C=C(O3)c(c2)ccc(O)c2)O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
0.2539 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2539 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8102 -0.5363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3665 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3665 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8102 0.7484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9228 -0.5363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4791 -0.2151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4791 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9228 0.7484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9228 -1.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0352 0.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6022 0.4209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1692 0.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1692 1.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6022 1.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0352 1.4030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3022 0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8102 -1.1784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7360 1.7302 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6147 0.6161 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.1801 0.0424 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5541 0.2858 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.9502 0.2923 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.3890 0.7313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0284 0.5017 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.5031 0.2828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8577 0.0094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1955 -0.3163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1608 0.5943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7360 0.2622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7360 -0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1608 -0.7340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5856 -0.4020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1608 1.0888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1608 -1.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5856 -1.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0104 -1.3982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0104 -0.7340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4353 -1.7303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2689 1.2346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6019 1.9797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
30 31 1 0 0 0 0
31 32 2 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
30 35 2 0 0 0 0
33 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
27 30 1 0 0 0 0
38 40 1 0 0 0 0
26 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 CH2OH
M SVB 1 45 -2.2689 1.2346
S SKP 8
ID FL3FAAGS0034
KNApSAcK_ID C00004170
NAME Apigenin 7-(4''-Z-p-coumarylglucoside)
CAS_RN -
FORMULA C30H26O12
EXACTMASS 578.1424262959999
AVERAGEMASS 578.5202400000001
SMILES O[C@H]([C@@H]1O)[C@@H](OC(C=Cc(c5)ccc(c5)O)=O)C(O[C@H]1Oc(c4)cc(c(c43)C(=O)C=C(O3)c(c2)ccc(O)c2)O)CO
M END