Mol:FL3FACGS0059
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 37 40 0 0 0 0 0 0 0 0999 V2000 | + | 37 40 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.0278 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0278 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0278 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0278 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4789 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4789 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9300 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9300 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9300 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9300 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4789 0.2003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4789 0.2003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3810 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3810 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8321 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8321 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8321 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8321 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3810 0.2003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3810 0.2003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5527 -1.2092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5527 -1.2092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2830 0.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2830 0.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7427 -0.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7427 -0.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2024 0.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2024 0.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2024 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2024 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7427 0.9965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7427 0.9965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2830 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2830 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5083 0.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5083 0.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4789 -1.3611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4789 -1.3611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7968 1.0742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7968 1.0742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7427 1.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7427 1.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1435 -0.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1435 -0.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1435 -0.6286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1435 -0.6286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7500 0.2136 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.7500 0.2136 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.7500 0.8993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7500 0.8993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4553 -0.1937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4553 -0.1937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0830 -0.4212 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.0830 -0.4212 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.5673 -1.1018 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.5673 -1.1018 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.8248 -0.8131 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.8248 -0.8131 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1084 -0.8053 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1084 -0.8053 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.6290 -0.2846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6290 -0.2846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2786 -0.6274 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.2786 -0.6274 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.7968 -0.8333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7968 -0.8333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3711 -1.1409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3711 -1.1409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3994 -1.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3994 -1.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4690 0.3526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4690 0.3526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4145 0.6783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4145 0.6783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 18 22 1 0 0 0 0 | + | 18 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 22 24 1 0 0 0 0 | + | 22 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 26 1 0 0 0 0 | + | 30 26 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 36 37 | + | M SAL 1 2 36 37 |
| − | M SBL 1 1 39 | + | M SBL 1 1 39 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 39 -3.469 0.3526 | + | M SVB 1 39 -3.469 0.3526 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACGS0059 | + | ID FL3FACGS0059 |
| − | KNApSAcK_ID C00004321 | + | KNApSAcK_ID C00004321 |
| − | NAME Luteolin 7-(2''-glucosyllactate) | + | NAME Luteolin 7-(2''-glucosyllactate) |
| − | CAS_RN 126394-93-2 | + | CAS_RN 126394-93-2 |
| − | FORMULA C24H24O13 | + | FORMULA C24H24O13 |
| − | EXACTMASS 520.121690854 | + | EXACTMASS 520.121690854 |
| − | AVERAGEMASS 520.43956 | + | AVERAGEMASS 520.43956 |
| − | SMILES c(c4)(O)c(cc(c4)C(=C3)Oc(c1)c(C3=O)c(cc(OC(C(C)O[C@@H]([C@@H](O)2)OC([C@H](O)[C@H](O)2)CO)=O)1)O)O | + | SMILES c(c4)(O)c(cc(c4)C(=C3)Oc(c1)c(C3=O)c(cc(OC(C(C)O[C@@H]([C@@H](O)2)OC([C@H](O)[C@H](O)2)CO)=O)1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
37 40 0 0 0 0 0 0 0 0999 V2000
1.0278 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0278 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4789 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9300 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9300 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4789 0.2003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3810 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8321 -0.5809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8321 -0.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3810 0.2003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5527 -1.2092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2830 0.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7427 -0.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2024 0.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2024 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7427 0.9965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2830 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5083 0.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4789 -1.3611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7968 1.0742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7427 1.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1435 -0.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1435 -0.6286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7500 0.2136 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.7500 0.8993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4553 -0.1937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0830 -0.4212 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.5673 -1.1018 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8248 -0.8131 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1084 -0.8053 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.6290 -0.2846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2786 -0.6274 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.7968 -0.8333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3711 -1.1409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3994 -1.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4690 0.3526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4145 0.6783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
16 21 1 0 0 0 0
18 22 1 0 0 0 0
22 23 2 0 0 0 0
22 24 1 0 0 0 0
24 25 1 0 0 0 0
24 26 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
27 33 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 26 1 0 0 0 0
32 36 1 0 0 0 0
36 37 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 36 37
M SBL 1 1 39
M SMT 1 CH2OH
M SVB 1 39 -3.469 0.3526
S SKP 8
ID FL3FACGS0059
KNApSAcK_ID C00004321
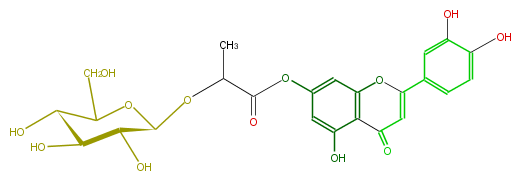
NAME Luteolin 7-(2''-glucosyllactate)
CAS_RN 126394-93-2
FORMULA C24H24O13
EXACTMASS 520.121690854
AVERAGEMASS 520.43956
SMILES c(c4)(O)c(cc(c4)C(=C3)Oc(c1)c(C3=O)c(cc(OC(C(C)O[C@@H]([C@@H](O)2)OC([C@H](O)[C@H](O)2)CO)=O)1)O)O
M END