Mol:FL3FADDS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8982 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8982 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8982 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8982 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1837 -2.0777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1837 -2.0777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5307 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5307 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5307 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5307 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1837 -0.4278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1837 -0.4278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2451 -2.0777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2451 -2.0777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9596 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9596 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9596 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9596 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2451 -0.4278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2451 -0.4278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2451 -2.7210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2451 -2.7210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1837 -2.9025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1837 -2.9025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8327 -0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8327 -0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5855 -0.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5855 -0.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3384 -0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3384 -0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3384 0.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3384 0.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5855 1.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5855 1.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8327 0.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8327 0.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2734 0.5929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2734 0.5929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6123 -0.4280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6123 -0.4280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5802 -0.3048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5802 -0.3048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9181 -1.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9181 -1.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2559 -0.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2559 -0.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3553 -0.7550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3553 -0.7550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9910 -0.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9910 -0.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7062 -0.5167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7062 -0.5167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2031 -0.6729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2031 -0.6729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6179 -1.1331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6179 -1.1331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6509 -1.0912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6509 -1.0912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1488 2.0281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1488 2.0281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2051 1.0354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2051 1.0354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4559 0.8482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4559 0.8482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1060 0.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1060 0.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0686 0.9979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0686 0.9979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6806 1.2602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6806 1.2602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8944 2.1348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8944 2.1348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8633 1.5439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8633 1.5439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3918 0.0801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3918 0.0801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9617 1.7554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9617 1.7554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5392 2.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5392 2.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7919 2.0699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7919 2.0699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1403 2.5758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1403 2.5758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3133 -0.0403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3133 -0.0403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2734 0.4074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2734 0.4074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 6 33 1 0 0 0 0 | + | 6 33 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 17 39 1 0 0 0 0 | + | 17 39 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -0.3762 -0.7471 | + | M SBV 1 44 -0.3762 -0.7471 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 45 0.1113 -0.8097 | + | M SBV 2 45 0.1113 -0.8097 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 ^ CH2OH | + | M SMT 3 ^ CH2OH |
| − | M SBV 3 48 0.6071 -0.4764 | + | M SBV 3 48 0.6071 -0.4764 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FADDS0002 | + | ID FL3FADDS0002 |
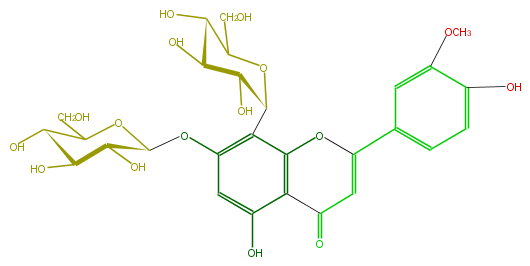
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
| − | SMILES C(C1O)(O)C(c(c24)c(OC(C(O)5)OC(CO)C(O)C5O)cc(c2C(=O)C=C(O4)c(c3)ccc(O)c3OC)O)OC(CO)C1O | + | SMILES C(C1O)(O)C(c(c24)c(OC(C(O)5)OC(CO)C(O)C5O)cc(c2C(=O)C=C(O4)c(c3)ccc(O)c3OC)O)OC(CO)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.8982 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8982 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1837 -2.0777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5307 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5307 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1837 -0.4278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2451 -2.0777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9596 -1.6653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9596 -0.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2451 -0.4278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2451 -2.7210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1837 -2.9025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8327 -0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5855 -0.7303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3384 -0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3384 0.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5855 1.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8327 0.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2734 0.5929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6123 -0.4280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5802 -0.3048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9181 -1.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2559 -0.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3553 -0.7550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9910 -0.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7062 -0.5167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2031 -0.6729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6179 -1.1331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6509 -1.0912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1488 2.0281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2051 1.0354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4559 0.8482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1060 0.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0686 0.9979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6806 1.2602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8944 2.1348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8633 1.5439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3918 0.0801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9617 1.7554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5392 2.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7919 2.0699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1403 2.5758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3133 -0.0403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2734 0.4074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 20 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
6 33 1 0 0 0 0
39 40 1 0 0 0 0
17 39 1 0 0 0 0
35 41 1 0 0 0 0
41 42 1 0 0 0 0
43 44 1 0 0 0 0
26 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -0.3762 -0.7471
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 CH2OH
M SBV 2 45 0.1113 -0.8097
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 48
M SMT 3 ^ CH2OH
M SBV 3 48 0.6071 -0.4764
S SKP 5
ID FL3FADDS0002
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES C(C1O)(O)C(c(c24)c(OC(C(O)5)OC(CO)C(O)C5O)cc(c2C(=O)C=C(O4)c(c3)ccc(O)c3OC)O)OC(CO)C1O
M END