Mol:FL3FAEGS0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.0350 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0350 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0350 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0350 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6422 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6422 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3196 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3196 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3196 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3196 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6422 -0.5428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6422 -0.5428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9969 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9969 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6741 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6741 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6741 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6741 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9969 -0.5428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9969 -0.5428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9969 -2.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9969 -2.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3512 -0.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3512 -0.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0415 -0.9415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0415 -0.9415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7319 -0.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7319 -0.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7319 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7319 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0415 0.6527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0415 0.6527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3512 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3512 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0415 1.4498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0415 1.4498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7109 -0.5436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7109 -0.5436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6422 -2.8874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6422 -2.8874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7375 -0.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7375 -0.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1182 -1.2049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1182 -1.2049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2262 -0.8580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2262 -0.8580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3655 -0.8487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3655 -0.8487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9910 -0.2232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9910 -0.2232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7714 -0.6350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7714 -0.6350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3860 -0.7201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3860 -0.7201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7922 -1.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7922 -1.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5488 -1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5488 -1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4488 0.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4488 0.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1323 0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1323 0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7674 0.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7674 0.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0127 -0.1896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0127 -0.1896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2523 0.7899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2523 0.7899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0127 1.7752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0127 1.7752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7674 2.2847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7674 2.2847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5281 1.3052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5281 1.3052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4290 2.8874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4290 2.8874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0127 2.4110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0127 2.4110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2260 1.7640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2260 1.7640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4447 0.2836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4447 0.2836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3720 0.7025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3720 0.7025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4447 0.0831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4447 0.0831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 31 30 1 0 0 0 0 | + | 31 30 1 0 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 32 1 1 0 0 0 | + | 37 32 1 1 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | 33 31 1 0 0 0 0 | + | 33 31 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 15 42 1 0 0 0 0 | + | 15 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 -0.6401 -0.4483 | + | M SBV 1 47 -0.6401 -0.4483 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAEGS0006 | + | ID FL3FAEGS0006 |
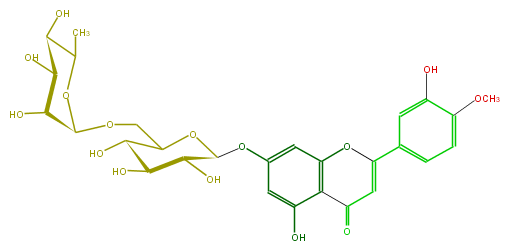
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES OC(C(O)4)C(COC(O5)C(C(C(O)C(C)5)O)O)OC(C(O)4)Oc(c3)cc(O1)c(c3O)C(C=C1c(c2)ccc(c2O)OC)=O | + | SMILES OC(C(O)4)C(COC(O5)C(C(C(O)C(C)5)O)O)OC(C(O)4)Oc(c3)cc(O1)c(c3O)C(C=C1c(c2)ccc(c2O)OC)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.0350 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0350 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6422 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3196 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3196 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6422 -0.5428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9969 -2.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6741 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6741 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9969 -0.5428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9969 -2.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3512 -0.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0415 -0.9415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7319 -0.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7319 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0415 0.6527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3512 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0415 1.4498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7109 -0.5436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6422 -2.8874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7375 -0.3873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1182 -1.2049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2262 -0.8580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3655 -0.8487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9910 -0.2232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7714 -0.6350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3860 -0.7201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7922 -1.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5488 -1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4488 0.0160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1323 0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.7674 0.3197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0127 -0.1896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2523 0.7899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0127 1.7752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7674 2.2847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5281 1.3052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4290 2.8874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0127 2.4110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.2260 1.7640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.4447 0.2836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3720 0.7025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.4447 0.0831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 19 1 0 0 0 0
26 30 1 0 0 0 0
31 30 1 0 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 32 1 1 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
37 40 1 0 0 0 0
32 41 1 0 0 0 0
33 31 1 0 0 0 0
42 43 1 0 0 0 0
15 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 -0.6401 -0.4483
S SKP 5
ID FL3FAEGS0006
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES OC(C(O)4)C(COC(O5)C(C(C(O)C(C)5)O)O)OC(C(O)4)Oc(c3)cc(O1)c(c3O)C(C=C1c(c2)ccc(c2O)OC)=O
M END