Mol:FL3FAGCS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0350 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0350 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0350 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0350 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4787 -1.8922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4787 -1.8922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0776 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0776 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0776 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0776 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4787 -0.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4787 -0.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6339 -1.8922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6339 -1.8922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1902 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1902 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1902 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1902 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6339 -0.6074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6339 -0.6074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6339 -2.3930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6339 -2.3930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5911 -0.6075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5911 -0.6075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8081 -0.5869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8081 -0.5869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3944 -0.9254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3944 -0.9254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9806 -0.5869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9806 -0.5869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9806 0.0900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9806 0.0900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3944 0.4284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3944 0.4284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8081 0.0900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8081 0.0900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5664 0.4282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5664 0.4282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2540 1.8708 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.2540 1.8708 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.8595 1.4120 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.8595 1.4120 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.6026 0.7514 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.6026 0.7514 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.5957 0.1139 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.5957 0.1139 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.1324 0.5771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1324 0.5771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4375 1.1551 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.4375 1.1551 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.4920 2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4920 2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8943 2.1272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8943 2.1272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2381 0.3728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2381 0.3728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4787 -2.5343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4787 -2.5343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9881 -1.9628 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.9881 -1.9628 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.6169 -2.4528 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.6169 -2.4528 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.0824 -2.2449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.0824 -2.2449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5667 -2.2394 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.5667 -2.2394 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9415 -1.8645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9415 -1.8645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4091 -2.1113 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.4091 -2.1113 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.5664 -2.1178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5664 -2.1178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1956 -2.4810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1956 -2.4810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7762 -2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7762 -2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5664 -0.9252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5664 -0.9252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3944 1.1049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3944 1.1049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0017 -1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0017 -1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4141 -0.5556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4141 -0.5556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3594 1.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3594 1.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8594 2.0211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8594 2.0211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 9 1 0 0 0 0 | + | 13 9 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 6 23 1 0 0 0 0 | + | 6 23 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 15 39 1 0 0 0 0 | + | 15 39 1 0 0 0 0 |
| − | 17 40 1 0 0 0 0 | + | 17 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 25 43 1 0 0 0 0 | + | 25 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 47 0.3594 1.1551 | + | M SVB 2 47 0.3594 1.1551 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 45 -2.5906 -1.0984 | + | M SVB 1 45 -2.5906 -1.0984 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAGCS0005 | + | ID FL3FAGCS0005 |
| − | KNApSAcK_ID C00006241 | + | KNApSAcK_ID C00006241 |
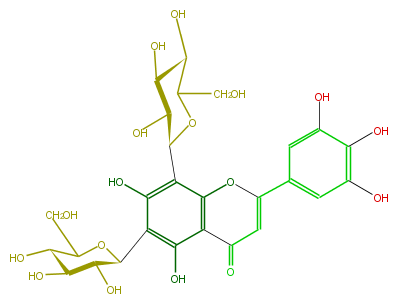
| − | NAME 6,8-Di-C-glucopyranosyltricetin | + | NAME 6,8-Di-C-glucopyranosyltricetin |
| − | CAS_RN 76491-11-7 | + | CAS_RN 76491-11-7 |
| − | FORMULA C27H30O17 | + | FORMULA C27H30O17 |
| − | EXACTMASS 626.148299534 | + | EXACTMASS 626.148299534 |
| − | AVERAGEMASS 626.5169000000001 | + | AVERAGEMASS 626.5169000000001 |
| − | SMILES c(c24)(c(O)c(c(O)c4[C@H](O5)[C@H]([C@H]([C@H](C5CO)O)O)O)[C@H](O3)[C@H]([C@H]([C@H](C(CO)3)O)O)O)C(C=C(O2)c(c1)cc(c(c(O)1)O)O)=O | + | SMILES c(c24)(c(O)c(c(O)c4[C@H](O5)[C@H]([C@H]([C@H](C5CO)O)O)O)[C@H](O3)[C@H]([C@H]([C@H](C(CO)3)O)O)O)C(C=C(O2)c(c1)cc(c(c(O)1)O)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.0350 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0350 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4787 -1.8922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0776 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0776 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4787 -0.6074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6339 -1.8922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1902 -1.5710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1902 -0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6339 -0.6074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6339 -2.3930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5911 -0.6075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8081 -0.5869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3944 -0.9254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9806 -0.5869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9806 0.0900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3944 0.4284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8081 0.0900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5664 0.4282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2540 1.8708 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.8595 1.4120 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6026 0.7514 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.5957 0.1139 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.1324 0.5771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4375 1.1551 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.4920 2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8943 2.1272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2381 0.3728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4787 -2.5343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9881 -1.9628 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.6169 -2.4528 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.0824 -2.2449 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5667 -2.2394 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9415 -1.8645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4091 -2.1113 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.5664 -2.1178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1956 -2.4810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7762 -2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5664 -0.9252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3944 1.1049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0017 -1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4141 -0.5556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3594 1.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8594 2.0211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
13 9 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
6 23 1 0 0 0 0
3 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 2 1 0 0 0 0
15 39 1 0 0 0 0
17 40 1 0 0 0 0
35 41 1 0 0 0 0
41 42 1 0 0 0 0
25 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 47
M SMT 2 CH2OH
M SVB 2 47 0.3594 1.1551
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 CH2OH
M SVB 1 45 -2.5906 -1.0984
S SKP 8
ID FL3FAGCS0005
KNApSAcK_ID C00006241
NAME 6,8-Di-C-glucopyranosyltricetin
CAS_RN 76491-11-7
FORMULA C27H30O17
EXACTMASS 626.148299534
AVERAGEMASS 626.5169000000001
SMILES c(c24)(c(O)c(c(O)c4[C@H](O5)[C@H]([C@H]([C@H](C5CO)O)O)O)[C@H](O3)[C@H]([C@H]([C@H](C(CO)3)O)O)O)C(C=C(O2)c(c1)cc(c(c(O)1)O)O)=O
M END