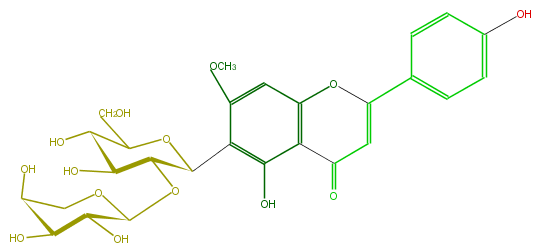
Mol:FL3FCACS0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8322 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8322 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8322 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8322 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1236 -0.7937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1236 -0.7937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5850 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5850 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5850 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5850 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1236 0.8429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1236 0.8429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2936 -0.7937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2936 -0.7937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0022 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0022 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0022 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0022 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2936 0.8429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2936 0.8429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2936 -1.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2936 -1.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1236 -1.6116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1236 -1.6116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8683 0.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8683 0.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6150 0.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6150 0.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3617 0.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3617 0.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3617 1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3617 1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6150 2.2673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6150 2.2673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8683 1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8683 1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1080 2.2671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1080 2.2671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7039 -0.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7039 -0.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1849 -0.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1849 -0.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4697 -0.6924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4697 -0.6924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6082 -0.9511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6082 -0.9511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1288 -0.2724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1288 -0.2724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8867 -0.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8867 -0.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3006 -0.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3006 -0.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0090 -0.9279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0090 -0.9279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9493 -1.3335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9493 -1.3335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0922 -1.5057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0922 -1.5057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4355 -2.2413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4355 -2.2413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7787 -1.8473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7787 -1.8473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8854 -1.9524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8854 -1.9524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5159 -1.3743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5159 -1.3743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2253 -1.7158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2253 -1.7158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0332 -0.8919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0332 -0.8919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1080 -2.3065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1080 -2.3065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1357 -2.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1357 -2.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2872 1.2218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2872 1.2218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9241 2.3249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9241 2.3249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5452 0.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5452 0.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6764 0.8401 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6764 0.8401 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 2 23 1 0 0 0 0 | + | 2 23 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 28 1 0 0 0 0 | + | 32 28 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 1 38 1 0 0 0 0 | + | 1 38 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 43 0.4550 -0.7881 | + | M SBV 1 43 0.4550 -0.7881 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 45 0.6585 -0.7401 | + | M SBV 2 45 0.6585 -0.7401 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FCACS0009 | + | ID FL3FCACS0009 |
| − | FORMULA C27H30O14 | + | FORMULA C27H30O14 |
| − | EXACTMASS 578.163555668 | + | EXACTMASS 578.163555668 |
| − | AVERAGEMASS 578.5187000000001 | + | AVERAGEMASS 578.5187000000001 |
| − | SMILES c(c21)(O)c(C(O4)C(OC(O5)C(C(O)C(O)C5)O)C(O)C(C(CO)4)O)c(cc1OC(c(c3)ccc(c3)O)=CC2=O)OC | + | SMILES c(c21)(O)c(C(O4)C(OC(O5)C(C(O)C(O)C5)O)C(O)C(C(CO)4)O)c(cc1OC(c(c3)ccc(c3)O)=CC2=O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-0.8322 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8322 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1236 -0.7937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5850 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5850 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1236 0.8429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2936 -0.7937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0022 -0.3846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0022 0.4337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2936 0.8429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2936 -1.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1236 -1.6116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8683 0.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6150 0.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3617 0.9741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3617 1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6150 2.2673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8683 1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1080 2.2671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7039 -0.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1849 -0.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4697 -0.6924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6082 -0.9511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1288 -0.2724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8867 -0.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3006 -0.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0090 -0.9279 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9493 -1.3335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0922 -1.5057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4355 -2.2413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7787 -1.8473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8854 -1.9524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5159 -1.3743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2253 -1.7158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0332 -0.8919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1080 -2.3065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1357 -2.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2872 1.2218 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9241 2.3249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5452 0.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6764 0.8401 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
2 23 1 0 0 0 0
22 28 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 28 1 0 0 0 0
38 39 1 0 0 0 0
1 38 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 43
M SMT 1 ^ OCH3
M SBV 1 43 0.4550 -0.7881
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 45
M SMT 2 ^ CH2OH
M SBV 2 45 0.6585 -0.7401
S SKP 5
ID FL3FCACS0009
FORMULA C27H30O14
EXACTMASS 578.163555668
AVERAGEMASS 578.5187000000001
SMILES c(c21)(O)c(C(O4)C(OC(O5)C(C(O)C(O)C5)O)C(O)C(C(CO)4)O)c(cc1OC(c(c3)ccc(c3)O)=CC2=O)OC
M END