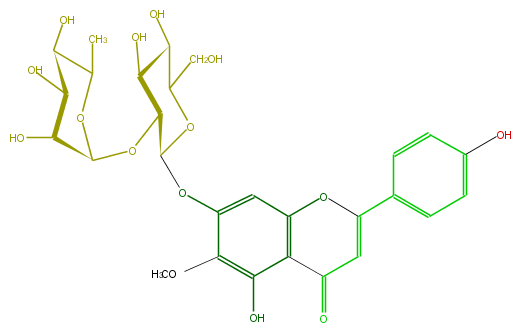
Mol:FL3FEAGS0031
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8474 -1.0402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8474 -1.0402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8474 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8474 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1464 -2.3183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1464 -2.3183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5544 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5544 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5544 -1.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5544 -1.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1464 -0.6995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1464 -0.6995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2554 -2.3183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2554 -2.3183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9564 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9564 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9564 -1.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9564 -1.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2554 -0.6995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2554 -0.6995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2555 -3.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2555 -3.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6571 -0.6997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6571 -0.6997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3715 -1.1121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3715 -1.1121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0858 -0.6997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0858 -0.6997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0858 0.1253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0858 0.1253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3715 0.5377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3715 0.5377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6571 0.1253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6571 0.1253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1464 -3.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1464 -3.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5641 -0.6262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5641 -0.6262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8000 0.5376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8000 0.5376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8321 2.3006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8321 2.3006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4358 1.6967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4358 1.6967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0130 0.9549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0130 0.9549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0130 0.1011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0130 0.1011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4092 0.7049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4092 0.7049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8321 1.4469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8321 1.4469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1361 2.9646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1361 2.9646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4980 2.4926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4980 2.4926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5671 0.2206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5671 0.2206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1417 0.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1417 0.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3607 0.0156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3607 0.0156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6086 0.8827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6086 0.8827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3607 1.7550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3607 1.7550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1417 2.2061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1417 2.2061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8940 1.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8940 1.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9357 2.8355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9357 2.8355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3647 2.4521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3647 2.4521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5744 1.8352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5744 1.8352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8000 0.4740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8000 0.4740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6263 -2.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6263 -2.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1545 -1.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1545 -1.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3421 2.0623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3421 2.0623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7205 3.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7205 3.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 1 1 0 0 0 0 | + | 19 1 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 19 24 1 0 0 0 0 | + | 19 24 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 30 39 1 0 0 0 0 | + | 30 39 1 0 0 0 0 |
| − | 31 29 1 0 0 0 0 | + | 31 29 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 2 40 1 0 0 0 0 | + | 2 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 26 42 1 0 0 0 0 | + | 26 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 45 0.7789 0.3325 | + | M SBV 1 45 0.7789 0.3325 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 47 -0.4900 -0.6154 | + | M SBV 2 47 -0.4900 -0.6154 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FEAGS0031 | + | ID FL3FEAGS0031 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES C(C(=O)2)=C(Oc(c3)c2c(O)c(OC)c3OC(C4OC(O5)C(C(C(C5C)O)O)O)OC(CO)C(C(O)4)O)c(c1)ccc(c1)O | + | SMILES C(C(=O)2)=C(Oc(c3)c2c(O)c(OC)c3OC(C4OC(O5)C(C(C(C5C)O)O)O)OC(CO)C(C(O)4)O)c(c1)ccc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.8474 -1.0402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8474 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1464 -2.3183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5544 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5544 -1.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1464 -0.6995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2554 -2.3183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9564 -1.9135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9564 -1.1042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2554 -0.6995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2555 -3.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6571 -0.6997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3715 -1.1121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0858 -0.6997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0858 0.1253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3715 0.5377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6571 0.1253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1464 -3.1259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5641 -0.6262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8000 0.5376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8321 2.3006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4358 1.6967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0130 0.9549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0130 0.1011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4092 0.7049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8321 1.4469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1361 2.9646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4980 2.4926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5671 0.2206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1417 0.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3607 0.0156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6086 0.8827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3607 1.7550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1417 2.2061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8940 1.3388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9357 2.8355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3647 2.4521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5744 1.8352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8000 0.4740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6263 -2.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1545 -1.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3421 2.0623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7205 3.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 1 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
19 24 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
35 38 1 0 0 0 0
30 39 1 0 0 0 0
31 29 1 0 0 0 0
40 41 1 0 0 0 0
2 40 1 0 0 0 0
42 43 1 0 0 0 0
26 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ OCH3
M SBV 1 45 0.7789 0.3325
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 CH2OH
M SBV 2 47 -0.4900 -0.6154
S SKP 5
ID FL3FEAGS0031
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES C(C(=O)2)=C(Oc(c3)c2c(O)c(OC)c3OC(C4OC(O5)C(C(C(C5C)O)O)O)OC(CO)C(C(O)4)O)c(c1)ccc(c1)O
M END