Mol:FL3FFCGS0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.1590 -1.0797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1590 -1.0797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1590 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1590 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5527 -2.1851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5527 -2.1851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9464 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9464 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9464 -1.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9464 -1.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5527 -0.7850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5527 -0.7850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3403 -2.1851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3403 -2.1851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7340 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7340 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7340 -1.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7340 -1.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3403 -0.7850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3403 -0.7850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3403 -2.9958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3403 -2.9958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1279 -0.7852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1279 -0.7852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4899 -1.1419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4899 -1.1419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1077 -0.7852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1077 -0.7852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1077 -0.0718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1077 -0.0718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4899 0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4899 0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1279 -0.0718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1279 -0.0718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4899 0.9876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4899 0.9876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5527 -2.8836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5527 -2.8836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6013 -0.8243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6013 -0.8243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5596 -0.1264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5596 -0.1264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1013 2.1925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1013 2.1925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4579 1.3465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4579 1.3465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7899 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7899 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3974 0.1474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3974 0.1474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1924 0.9121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1924 0.9121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8609 1.3326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8609 1.3326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8262 2.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8262 2.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8632 1.9282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8632 1.9282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7916 -0.1361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7916 -0.1361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7098 0.3686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7098 0.3686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0107 0.3443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0107 0.3443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2887 -0.0725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2887 -0.0725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0975 -0.2749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0975 -0.2749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6812 -0.8765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6812 -0.8765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4033 -0.4597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4033 -0.4597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5946 -0.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5946 -0.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3469 0.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3469 0.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1662 0.0728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1662 0.0728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6700 -0.9236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6700 -0.9236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4883 -1.3135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4883 -1.3135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8632 -2.1175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8632 -2.1175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7202 -1.7570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7202 -1.7570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6983 2.2279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6983 2.2279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6393 2.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6393 2.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2548 2.9958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2548 2.9958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 6 21 1 0 0 0 0 | + | 6 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 21 1 0 0 0 0 | + | 25 21 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 33 31 1 0 0 0 0 | + | 33 31 1 0 0 0 0 |
| − | 31 15 1 0 0 0 0 | + | 31 15 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 27 44 1 0 0 0 0 | + | 27 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 41 42 43 | + | M SAL 1 3 41 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 47 -0.8071 0.4370 | + | M SBV 1 47 -0.8071 0.4370 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 44 45 46 | + | M SAL 2 3 44 45 46 |
| − | M SBL 2 1 50 | + | M SBL 2 1 50 |
| − | M SMT 2 COOH | + | M SMT 2 COOH |
| − | M SBV 2 50 -0.1627 -0.8952 | + | M SBV 2 50 -0.1627 -0.8952 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FFCGS0007 | + | ID FL3FFCGS0007 |
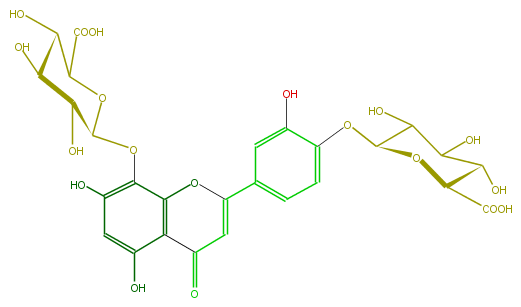
| − | FORMULA C27H26O19 | + | FORMULA C27H26O19 |
| − | EXACTMASS 654.1068286499999 | + | EXACTMASS 654.1068286499999 |
| − | AVERAGEMASS 654.4839400000001 | + | AVERAGEMASS 654.4839400000001 |
| − | SMILES c(c1O)c(C(=C4)Oc(c3OC(O5)C(O)C(C(C5C(O)=O)O)O)c(C4=O)c(O)cc(O)3)ccc1OC(C(O)2)OC(C(O)=O)C(O)C(O)2 | + | SMILES c(c1O)c(C(=C4)Oc(c3OC(O5)C(O)C(C(C5C(O)=O)O)O)c(C4=O)c(O)cc(O)3)ccc1OC(C(O)2)OC(C(O)=O)C(O)C(O)2 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-3.1590 -1.0797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1590 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5527 -2.1851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9464 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9464 -1.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5527 -0.7850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3403 -2.1851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7340 -1.8351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7340 -1.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3403 -0.7850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3403 -2.9958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1279 -0.7852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4899 -1.1419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1077 -0.7852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1077 -0.0718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4899 0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1279 -0.0718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4899 0.9876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5527 -2.8836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6013 -0.8243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5596 -0.1264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1013 2.1925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4579 1.3465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7899 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3974 0.1474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1924 0.9121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8609 1.3326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8262 2.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8632 1.9282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7916 -0.1361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7098 0.3686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0107 0.3443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2887 -0.0725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0975 -0.2749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6812 -0.8765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4033 -0.4597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5946 -0.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3469 0.6577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1662 0.0728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6700 -0.9236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4883 -1.3135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8632 -2.1175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7202 -1.7570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6983 2.2279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6393 2.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2548 2.9958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
3 19 1 0 0 0 0
1 20 1 0 0 0 0
6 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 21 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
33 31 1 0 0 0 0
31 15 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
35 41 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
27 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 41 42 43
M SBL 1 1 47
M SMT 1 COOH
M SBV 1 47 -0.8071 0.4370
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 44 45 46
M SBL 2 1 50
M SMT 2 COOH
M SBV 2 50 -0.1627 -0.8952
S SKP 5
ID FL3FFCGS0007
FORMULA C27H26O19
EXACTMASS 654.1068286499999
AVERAGEMASS 654.4839400000001
SMILES c(c1O)c(C(=C4)Oc(c3OC(O5)C(O)C(C(C5C(O)=O)O)O)c(C4=O)c(O)cc(O)3)ccc1OC(C(O)2)OC(C(O)=O)C(O)C(O)2
M END