Mol:FL3FFGCS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.9168 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9168 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9168 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9168 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3605 -0.9738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3605 -0.9738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1958 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1958 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1958 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1958 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3605 0.3109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3605 0.3109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7521 -0.9738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7521 -0.9738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3084 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3084 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3084 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3084 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7521 0.3109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7521 0.3109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7521 -1.4747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7521 -1.4747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3605 -1.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3605 -1.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4701 -0.8972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.4701 -0.8972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.9545 -1.4747 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.9545 -1.4747 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.4389 -1.1653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4389 -1.1653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.7376 -1.2478 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.7376 -1.2478 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2326 -0.7940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2326 -0.7940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7895 -1.0622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.7895 -1.0622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.1697 -1.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1697 -1.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7655 -1.4747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7655 -1.4747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4729 0.3108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4729 0.3108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8690 0.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8690 0.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3978 0.0285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3978 0.0285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9265 0.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9265 0.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9265 0.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9265 0.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3978 1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3978 1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8690 0.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8690 0.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0639 -1.8147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0639 -1.8147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7925 1.4443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7925 1.4443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6586 1.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6586 1.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0824 0.8839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0824 0.8839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3542 1.7835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3542 1.7835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6621 1.8147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6621 1.8147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0858 2.7205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0858 2.7205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7925 -0.1662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7925 -0.1662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6586 -0.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6586 -0.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9906 -0.1751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9906 -0.1751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9457 0.1211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9457 0.1211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 2 16 1 0 0 0 0 | + | 2 16 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 9 22 1 0 0 0 0 | + | 9 22 1 0 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 6 31 1 0 0 0 0 | + | 6 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 26 33 1 0 0 0 0 | + | 26 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 24 35 1 0 0 0 0 | + | 24 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 18 37 1 0 0 0 0 | + | 18 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 37 38 | + | M SAL 5 2 37 38 |
| − | M SBL 5 1 40 | + | M SBL 5 1 40 |
| − | M SMT 5 CH2OH | + | M SMT 5 CH2OH |
| − | M SVB 5 40 -2.9906 -0.1751 | + | M SVB 5 40 -2.9906 -0.1751 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 35 36 | + | M SAL 4 2 35 36 |
| − | M SBL 4 1 38 | + | M SBL 4 1 38 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 38 3.098 0.1275 | + | M SVB 4 38 3.098 0.1275 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 33 34 | + | M SAL 3 2 33 34 |
| − | M SBL 3 1 36 | + | M SBL 3 1 36 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 36 2.6621 1.8147 | + | M SVB 3 36 2.6621 1.8147 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 31 32 | + | M SAL 2 2 31 32 |
| − | M SBL 2 1 34 | + | M SBL 2 1 34 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 34 -0.0824 0.8839 | + | M SVB 2 34 -0.0824 0.8839 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 29 30 | + | M SAL 1 2 29 30 |
| − | M SBL 1 1 32 | + | M SBL 1 1 32 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 32 3.4552 1.2496 | + | M SVB 1 32 3.4552 1.2496 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FFGCS0001 | + | ID FL3FFGCS0001 |
| − | KNApSAcK_ID C00006371 | + | KNApSAcK_ID C00006371 |
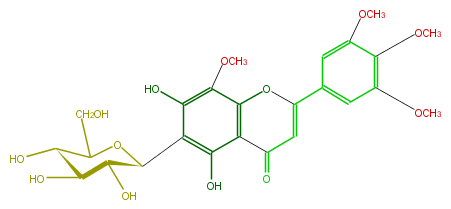
| − | NAME 5,7-Dihydroxy-8,3',4',5'-tetramethoxyflavone 6-C-glucoside | + | NAME 5,7-Dihydroxy-8,3',4',5'-tetramethoxyflavone 6-C-glucoside |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C25H28O13 | + | FORMULA C25H28O13 |
| − | EXACTMASS 536.152990982 | + | EXACTMASS 536.152990982 |
| − | AVERAGEMASS 536.48202 | + | AVERAGEMASS 536.48202 |
| − | SMILES c(c3O)(C(=O)1)c(c(c(c3[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)O)OC)OC(c(c2)cc(OC)c(OC)c2OC)=C1 | + | SMILES c(c3O)(C(=O)1)c(c(c(c3[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)O)OC)OC(c(c2)cc(OC)c(OC)c2OC)=C1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-0.9168 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9168 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3605 -0.9738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1958 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1958 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3605 0.3109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7521 -0.9738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3084 -0.6526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3084 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7521 0.3109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7521 -1.4747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3605 -1.6160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4701 -0.8972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.9545 -1.4747 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.4389 -1.1653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.7376 -1.2478 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2326 -0.7940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7895 -1.0622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.1697 -1.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7655 -1.4747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4729 0.3108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8690 0.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3978 0.0285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9265 0.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9265 0.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3978 1.2496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8690 0.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0639 -1.8147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7925 1.4443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6586 1.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0824 0.8839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3542 1.7835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6621 1.8147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0858 2.7205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7925 -0.1662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6586 -0.6662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9906 -0.1751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9457 0.1211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
2 16 1 0 0 0 0
1 21 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 2 0 0 0 0
25 26 1 0 0 0 0
26 27 2 0 0 0 0
27 22 1 0 0 0 0
9 22 1 0 0 0 0
15 28 1 0 0 0 0
25 29 1 0 0 0 0
29 30 1 0 0 0 0
6 31 1 0 0 0 0
31 32 1 0 0 0 0
26 33 1 0 0 0 0
33 34 1 0 0 0 0
24 35 1 0 0 0 0
35 36 1 0 0 0 0
18 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 37 38
M SBL 5 1 40
M SMT 5 CH2OH
M SVB 5 40 -2.9906 -0.1751
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 35 36
M SBL 4 1 38
M SMT 4 OCH3
M SVB 4 38 3.098 0.1275
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 33 34
M SBL 3 1 36
M SMT 3 OCH3
M SVB 3 36 2.6621 1.8147
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 31 32
M SBL 2 1 34
M SMT 2 OCH3
M SVB 2 34 -0.0824 0.8839
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 29 30
M SBL 1 1 32
M SMT 1 OCH3
M SVB 1 32 3.4552 1.2496
S SKP 8
ID FL3FFGCS0001
KNApSAcK_ID C00006371
NAME 5,7-Dihydroxy-8,3',4',5'-tetramethoxyflavone 6-C-glucoside
CAS_RN -
FORMULA C25H28O13
EXACTMASS 536.152990982
AVERAGEMASS 536.48202
SMILES c(c3O)(C(=O)1)c(c(c(c3[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)O)OC)OC(c(c2)cc(OC)c(OC)c2OC)=C1
M END