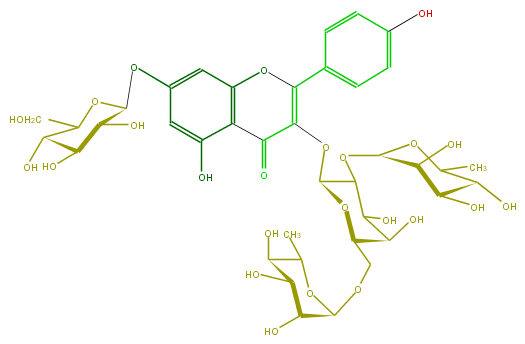
Mol:FL5FAAGA0027
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 63 69 0 0 0 0 0 0 0 0999 V2000 | + | 63 69 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.9687 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9687 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9687 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9687 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2542 0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2542 0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5397 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5397 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5397 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5397 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2542 2.3483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2542 2.3483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1747 0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1747 0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8892 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8892 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8892 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8892 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1747 2.3483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1747 2.3483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1747 -0.0608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1747 -0.0608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6032 2.3834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6032 2.3834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3314 1.9629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3314 1.9629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0595 2.3834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0595 2.3834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0595 3.2241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0595 3.2241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3314 3.6446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3314 3.6446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6032 3.2241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6032 3.2241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2542 -0.1263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2542 -0.1263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8389 3.6741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8389 3.6741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6126 0.5849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6126 0.5849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8221 2.4286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8221 2.4286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2867 -0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2867 -0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4663 -0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4663 -0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0559 -0.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0559 -0.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2576 -1.6075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2576 -1.6075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0780 -1.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0780 -1.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4884 -1.0370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4884 -1.0370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0164 0.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0164 0.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9975 -1.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9975 -1.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6122 -1.0903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6122 -1.0903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6352 -2.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6352 -2.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3651 -2.7037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3651 -2.7037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0325 -3.3358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0325 -3.3358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8113 -3.3358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8113 -3.3358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2514 -2.7942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2514 -2.7942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0601 -2.0347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0601 -2.0347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2812 -2.0347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2812 -2.0347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8410 -2.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8410 -2.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2812 -1.3990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2812 -1.3990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7127 -1.4329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7127 -1.4329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0090 -2.3485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0090 -2.3485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4465 -3.6741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4465 -3.6741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8983 0.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8983 0.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1126 0.4516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1126 0.4516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6147 1.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6147 1.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9991 1.4364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9991 1.4364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7093 1.6267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7093 1.6267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0998 1.0059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0998 1.0059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2787 0.1238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2787 0.1238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6754 0.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6754 0.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8206 1.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8206 1.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6513 0.6563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6513 0.6563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2646 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2646 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1035 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1035 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7691 -0.7824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7691 -0.7824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2287 -0.5592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2287 -0.5592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7305 0.1246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7305 0.1246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9885 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9885 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5074 0.6487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5074 0.6487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8373 0.3617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8373 0.3617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0421 -0.8145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0421 -0.8145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8925 1.2183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8925 1.2183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7691 1.9538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7691 1.9538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 38 33 1 1 0 0 0 | + | 38 33 1 1 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 33 42 1 0 0 0 0 | + | 33 42 1 0 0 0 0 |
| − | 34 32 1 0 0 0 0 | + | 34 32 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 21 1 0 0 0 0 | + | 46 21 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 54 56 1 1 0 0 0 | + | 54 56 1 1 0 0 0 |
| − | 56 57 1 1 0 0 0 | + | 56 57 1 1 0 0 0 |
| − | 53 58 1 0 0 0 0 | + | 53 58 1 0 0 0 0 |
| − | 57 59 1 0 0 0 0 | + | 57 59 1 0 0 0 0 |
| − | 28 60 1 0 0 0 0 | + | 28 60 1 0 0 0 0 |
| − | 60 57 1 1 0 0 0 | + | 60 57 1 1 0 0 0 |
| − | 60 52 1 0 0 0 0 | + | 60 52 1 0 0 0 0 |
| − | 56 61 1 0 0 0 0 | + | 56 61 1 0 0 0 0 |
| − | 62 63 1 0 0 0 0 | + | 62 63 1 0 0 0 0 |
| − | 48 62 1 0 0 0 0 | + | 48 62 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 62 63 | + | M SAL 1 2 62 63 |
| − | M SBL 1 1 69 | + | M SBL 1 1 69 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 69 0.7927 -0.2124 | + | M SBV 1 69 0.7927 -0.2124 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGA0027 | + | ID FL5FAAGA0027 |
| − | FORMULA C39H50O24 | + | FORMULA C39H50O24 |
| − | EXACTMASS 902.269202528 | + | EXACTMASS 902.269202528 |
| − | AVERAGEMASS 902.7999 | + | AVERAGEMASS 902.7999 |
| − | SMILES C(C1Oc(c7)cc(c(c7O)5)OC(c(c6)ccc(c6)O)=C(C(=O)5)OC(C3OC(C(O)4)OC(C)C(O)C4O)OC(C(C3O)O)COC(C2O)OC(C(C2O)O)C)(O)C(O)C(O)C(CO)O1 | + | SMILES C(C1Oc(c7)cc(c(c7O)5)OC(c(c6)ccc(c6)O)=C(C(=O)5)OC(C3OC(C(O)4)OC(C)C(O)C4O)OC(C(C3O)O)COC(C2O)OC(C(C2O)O)C)(O)C(O)C(O)C(CO)O1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
63 69 0 0 0 0 0 0 0 0999 V2000
-1.9687 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9687 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2542 0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5397 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5397 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2542 2.3483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1747 0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8892 1.1109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8892 1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1747 2.3483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1747 -0.0608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6032 2.3834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3314 1.9629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0595 2.3834 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0595 3.2241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3314 3.6446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6032 3.2241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2542 -0.1263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8389 3.6741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6126 0.5849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8221 2.4286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2867 -0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4663 -0.2370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0559 -0.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2576 -1.6075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0780 -1.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4884 -1.0370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0164 0.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9975 -1.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6122 -1.0903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6352 -2.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3651 -2.7037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0325 -3.3358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8113 -3.3358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2514 -2.7942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0601 -2.0347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2812 -2.0347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8410 -2.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2812 -1.3990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7127 -1.4329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0090 -2.3485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4465 -3.6741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8983 0.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1126 0.4516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6147 1.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9991 1.4364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7093 1.6267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0998 1.0059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2787 0.1238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6754 0.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8206 1.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6513 0.6563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2646 0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1035 -0.2668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7691 -0.7824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2287 -0.5592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7305 0.1246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9885 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5074 0.6487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8373 0.3617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0421 -0.8145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8925 1.2183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7691 1.9538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
1 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
23 20 1 0 0 0 0
31 32 1 0 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 1 0 0 0
38 33 1 1 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
38 41 1 0 0 0 0
33 42 1 0 0 0 0
34 32 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 21 1 0 0 0 0
52 53 1 0 0 0 0
53 54 1 0 0 0 0
54 55 1 0 0 0 0
54 56 1 1 0 0 0
56 57 1 1 0 0 0
53 58 1 0 0 0 0
57 59 1 0 0 0 0
28 60 1 0 0 0 0
60 57 1 1 0 0 0
60 52 1 0 0 0 0
56 61 1 0 0 0 0
62 63 1 0 0 0 0
48 62 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 62 63
M SBL 1 1 69
M SMT 1 ^ CH2OH
M SBV 1 69 0.7927 -0.2124
S SKP 5
ID FL5FAAGA0027
FORMULA C39H50O24
EXACTMASS 902.269202528
AVERAGEMASS 902.7999
SMILES C(C1Oc(c7)cc(c(c7O)5)OC(c(c6)ccc(c6)O)=C(C(=O)5)OC(C3OC(C(O)4)OC(C)C(O)C4O)OC(C(C3O)O)COC(C2O)OC(C(C2O)O)C)(O)C(O)C(O)C(CO)O1
M END