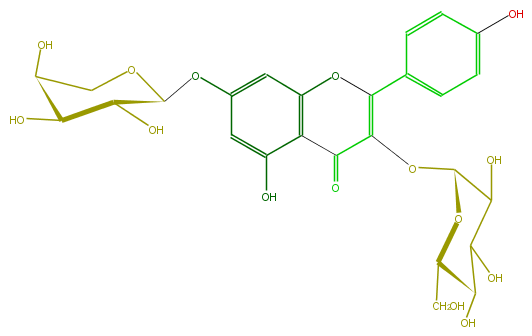
Mol:FL5FAAGL0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5330 -0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5330 -0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2641 -0.7215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2641 -0.7215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5409 0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5409 0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0206 0.6592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0206 0.6592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7765 0.5187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7765 0.5187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0533 -0.2420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0533 -0.2420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2975 1.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2975 1.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2228 2.0398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2228 2.0398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0199 1.8993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0199 1.8993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2968 1.1386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2968 1.1386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9188 1.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9188 1.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5400 2.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5400 2.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2578 3.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2578 3.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7881 3.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7881 3.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6006 3.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6006 3.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8828 3.0078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8828 3.0078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3525 2.3759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3525 2.3759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3377 0.1797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3377 0.1797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8098 -1.6223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8098 -1.6223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1307 4.4148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1307 4.4148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2626 2.9810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2626 2.9810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6449 4.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6449 4.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1563 3.7955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1563 3.7955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0957 4.0639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0957 4.0639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0407 3.7955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0407 3.7955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5293 4.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5293 4.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5899 4.3731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5899 4.3731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1785 4.4795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1785 4.4795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1430 4.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1430 4.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1307 4.5370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1307 4.5370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2242 -4.7750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2242 -4.7750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5559 -4.1096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5559 -4.1096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0150 -3.1532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0150 -3.1532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1734 -2.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1734 -2.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7057 -2.9007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7057 -2.9007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1381 -3.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1381 -3.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8756 -4.7668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8756 -4.7668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6570 -4.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6570 -4.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3956 -2.2833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3956 -2.2833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8975 3.9146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8975 3.9146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0779 2.6675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0779 2.6675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.8568 -0.1192 | + | M SBV 1 45 -0.8568 -0.1192 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0012 | + | ID FL5FAAGL0012 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(O)c(C(=O)3)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2)O | + | SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(O)c(C(=O)3)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-0.5330 -0.8620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2641 -0.7215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5409 0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0206 0.6592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7765 0.5187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0533 -0.2420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2975 1.4198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2228 2.0398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0199 1.8993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2968 1.1386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9188 1.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5400 2.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2578 3.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7881 3.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6006 3.7830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8828 3.0078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3525 2.3759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3377 0.1797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8098 -1.6223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1307 4.4148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2626 2.9810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6449 4.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1563 3.7955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0957 4.0639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0407 3.7955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5293 4.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5899 4.3731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1785 4.4795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1430 4.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1307 4.5370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2242 -4.7750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5559 -4.1096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0150 -3.1532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1734 -2.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7057 -2.9007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1381 -3.6318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8756 -4.7668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6570 -4.9178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3956 -2.2833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8975 3.9146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0779 2.6675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
8 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 19 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.8568 -0.1192
S SKP 5
ID FL5FAAGL0012
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES OC(C(O)5)C(OCC5O)Oc(c1)cc(O)c(C(=O)3)c1OC(=C3OC(C(O)4)OC(C(C(O)4)O)CO)c(c2)ccc(c2)O
M END