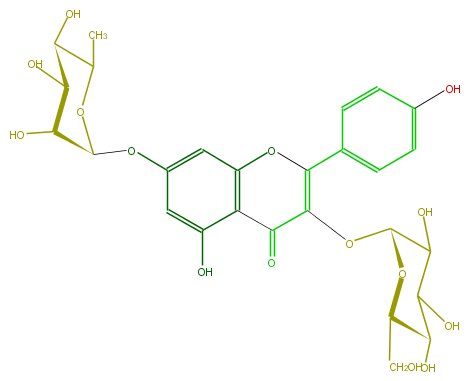
Mol:FL5FAAGL0016
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3613 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3613 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3613 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3613 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6602 -0.2719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6602 -0.2719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0408 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0408 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0408 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0408 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6602 1.3473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6602 1.3473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7419 -0.2719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7419 -0.2719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4429 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4429 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4429 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4429 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7419 1.3473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7419 1.3473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7419 -0.9030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7419 -0.9030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1437 1.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1437 1.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8583 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8583 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5728 1.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5728 1.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5728 2.1721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5728 2.1721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8583 2.5847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8583 2.5847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1437 2.1721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1437 2.1721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6602 -1.0811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6602 -1.0811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0621 1.3470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0621 1.3470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2872 2.5846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2872 2.5846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2856 -0.5086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2856 -0.5086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1048 -1.9946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1048 -1.9946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8965 -2.4517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8965 -2.4517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6454 -1.5727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6454 -1.5727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8965 -0.6884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8965 -0.6884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1048 -0.2312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1048 -0.2312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3560 -1.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3560 -1.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7862 -3.0021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7862 -3.0021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2633 -2.1491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2633 -2.1491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6198 1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6198 1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8280 1.2502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8280 1.2502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0792 2.1292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0792 2.1292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8280 3.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8280 3.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6198 3.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6198 3.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3685 2.5917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3685 2.5917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3141 4.0865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3141 4.0865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8483 3.6687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8483 3.6687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0588 3.0905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0588 3.0905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2872 1.6711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2872 1.6711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7427 0.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7427 0.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0727 -2.9819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0727 -2.9819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6795 -4.0865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6795 -4.0865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 24 29 1 0 0 0 0 | + | 24 29 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 30 39 1 0 0 0 0 | + | 30 39 1 0 0 0 0 |
| − | 31 19 1 0 0 0 0 | + | 31 19 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 22 41 1 0 0 0 0 | + | 22 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 46 0.0321 0.9872 | + | M SBV 1 46 0.0321 0.9872 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0016 | + | ID FL5FAAGL0016 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES c(c25)(O)cc(cc2OC(=C(C5=O)OC(C(O)4)OC(C(C(O)4)O)CO)c(c3)ccc(c3)O)OC(C(O)1)OC(C)C(O)C1O | + | SMILES c(c25)(O)cc(cc2OC(=C(C5=O)OC(C(O)4)OC(C(C(O)4)O)CO)c(c3)ccc(c3)O)OC(C(O)1)OC(C)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-1.3613 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3613 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6602 -0.2719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0408 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0408 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6602 1.3473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7419 -0.2719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4429 0.1329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4429 0.9425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7419 1.3473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7419 -0.9030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1437 1.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8583 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5728 1.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5728 2.1721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8583 2.5847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1437 2.1721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6602 -1.0811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0621 1.3470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2872 2.5846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2856 -0.5086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1048 -1.9946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8965 -2.4517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6454 -1.5727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8965 -0.6884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1048 -0.2312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3560 -1.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7862 -3.0021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2633 -2.1491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6198 1.7073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8280 1.2502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0792 2.1292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8280 3.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6198 3.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3685 2.5917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3141 4.0865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8483 3.6687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0588 3.0905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2872 1.6711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7427 0.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0727 -2.9819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6795 -4.0865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
23 28 1 0 0 0 0
24 29 1 0 0 0 0
26 21 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
35 38 1 0 0 0 0
30 39 1 0 0 0 0
31 19 1 0 0 0 0
25 40 1 0 0 0 0
41 42 1 0 0 0 0
22 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^ CH2OH
M SBV 1 46 0.0321 0.9872
S SKP 5
ID FL5FAAGL0016
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES c(c25)(O)cc(cc2OC(=C(C5=O)OC(C(O)4)OC(C(C(O)4)O)CO)c(c3)ccc(c3)O)OC(C(O)1)OC(C)C(O)C1O
M END