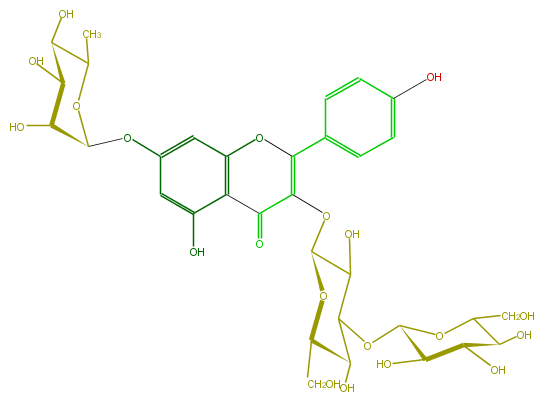
Mol:FL5FAAGL0038
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3376 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3376 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3376 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3376 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6365 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6365 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9355 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9355 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9355 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9355 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6365 1.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6365 1.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2345 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2345 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4665 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4665 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4665 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4665 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2345 1.3338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2345 1.3338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2345 -0.9162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2345 -0.9162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1673 1.3337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1673 1.3337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8818 0.9212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8818 0.9212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5963 1.3337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5963 1.3337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5963 2.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5963 2.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8818 2.5711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8818 2.5711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1673 2.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1673 2.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0383 1.3337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0383 1.3337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1804 -0.3261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1804 -0.3261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6365 -1.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6365 -1.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3951 2.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3951 2.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6602 1.5863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6602 1.5863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8686 1.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8686 1.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1197 2.0082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1197 2.0082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8686 2.8924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8686 2.8924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6602 3.3496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6602 3.3496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4091 2.4705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4091 2.4705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4144 3.9701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4144 3.9701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9194 3.5369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9194 3.5369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0579 2.9693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0579 2.9693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3070 1.5707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3070 1.5707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6979 -1.5777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6979 -1.5777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8518 -1.0893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8518 -1.0893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1204 -2.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1204 -2.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8518 -2.9737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8518 -2.9737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6979 -3.4624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6979 -3.4624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4296 -2.5230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4296 -2.5230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6638 -0.7145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6638 -0.7145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0596 -3.0761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0596 -3.0761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5449 -3.9701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5449 -3.9701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7246 -2.6554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7246 -2.6554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2821 -3.3914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2821 -3.3914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0849 -3.0791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0849 -3.0791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8595 -3.0708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8595 -3.0708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2966 -2.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2966 -2.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5943 -2.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5943 -2.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4763 -3.4763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4763 -3.4763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7525 -3.5846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7525 -3.5846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3070 -2.8481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3070 -2.8481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7594 -3.8849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7594 -3.8849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5005 -3.9093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5005 -3.9093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9851 -2.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9851 -2.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7488 -1.2123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7488 -1.2123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 23 18 1 0 0 0 0 | + | 23 18 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 33 19 1 0 0 0 0 | + | 33 19 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 42 47 1 0 0 0 0 | + | 42 47 1 0 0 0 0 |
| − | 43 48 1 0 0 0 0 | + | 43 48 1 0 0 0 0 |
| − | 41 39 1 0 0 0 0 | + | 41 39 1 0 0 0 0 |
| − | 44 49 1 0 0 0 0 | + | 44 49 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 35 50 1 0 0 0 0 | + | 35 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 45 52 1 0 0 0 0 | + | 45 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 56 0.0924 0.9112 | + | M SBV 1 56 0.0924 0.9112 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 58 -0.6884 -0.0576 | + | M SBV 2 58 -0.6884 -0.0576 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0038 | + | ID FL5FAAGL0038 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES C(C1O)(O)C(C(C)OC1Oc(c2)cc(O3)c(C(C(OC(C5O)OC(CO)C(C5OC(O6)C(C(O)C(C6CO)O)O)O)=C3c(c4)ccc(O)c4)=O)c2O)O | + | SMILES C(C1O)(O)C(C(C)OC1Oc(c2)cc(O3)c(C(C(OC(C5O)OC(CO)C(C5OC(O6)C(C(O)C(C6CO)O)O)O)=C3c(c4)ccc(O)c4)=O)c2O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-2.3376 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3376 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6365 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9355 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9355 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6365 1.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2345 -0.2851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4665 0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4665 0.9290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2345 1.3338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2345 -0.9162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1673 1.3337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8818 0.9212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5963 1.3337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5963 2.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8818 2.5711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1673 2.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0383 1.3337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1804 -0.3261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6365 -1.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3951 2.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6602 1.5863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8686 1.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1197 2.0082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8686 2.8924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6602 3.3496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4091 2.4705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4144 3.9701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9194 3.5369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0579 2.9693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3070 1.5707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6979 -1.5777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8518 -1.0893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1204 -2.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8518 -2.9737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6979 -3.4624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4296 -2.5230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6638 -0.7145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0596 -3.0761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5449 -3.9701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7246 -2.6554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2821 -3.3914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0849 -3.0791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8595 -3.0708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2966 -2.5077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5943 -2.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4763 -3.4763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7525 -3.5846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3070 -2.8481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7594 -3.8849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5005 -3.9093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9851 -2.4501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7488 -1.2123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
27 30 1 0 0 0 0
22 31 1 0 0 0 0
23 18 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
33 19 1 0 0 0 0
41 42 1 1 0 0 0
42 43 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
42 47 1 0 0 0 0
43 48 1 0 0 0 0
41 39 1 0 0 0 0
44 49 1 0 0 0 0
50 51 1 0 0 0 0
35 50 1 0 0 0 0
52 53 1 0 0 0 0
45 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 ^ CH2OH
M SBV 1 56 0.0924 0.9112
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 CH2OH
M SBV 2 58 -0.6884 -0.0576
S SKP 5
ID FL5FAAGL0038
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES C(C1O)(O)C(C(C)OC1Oc(c2)cc(O3)c(C(C(OC(C5O)OC(CO)C(C5OC(O6)C(C(O)C(C6CO)O)O)O)=C3c(c4)ccc(O)c4)=O)c2O)O
M END