Mol:FL5FABGI0017
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 51 55 0 0 0 0 0 0 0 0999 V2000 | + | 51 55 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.7150 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7150 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7150 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7150 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1587 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1587 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6024 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6024 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6024 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6024 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1587 0.9940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1587 0.9940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0461 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0461 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4898 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4898 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4898 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4898 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0461 0.9940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0461 0.9940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0461 -0.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0461 -0.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7893 1.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7893 1.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2223 0.7903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2223 0.7903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3447 1.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3447 1.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3447 1.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3447 1.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2223 2.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2223 2.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7893 1.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7893 1.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3795 1.0564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3795 1.0564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1587 -0.8535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1587 -0.8535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9058 -0.3585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9058 -0.3585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1469 -1.4622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1469 -1.4622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6823 -1.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6823 -1.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5124 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5124 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6823 -0.5788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6823 -0.5788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1469 -0.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1469 -0.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3167 -0.8641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3167 -0.8641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9733 -0.8641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9733 -0.8641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2844 -1.9755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2844 -1.9755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5531 -2.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5531 -2.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0410 -1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0410 -1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3391 -2.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3391 -2.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7516 -2.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7516 -2.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3391 -2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3391 -2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0673 -0.5587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0673 -0.5587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4690 -1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4690 -1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0475 -0.8640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0475 -0.8640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6057 -0.8579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6057 -0.8579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2001 -0.4523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2001 -0.4523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6940 -0.7194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6940 -0.7194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8428 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8428 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3789 -1.4204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3789 -1.4204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0612 -0.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0612 -0.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1587 1.6361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1587 1.6361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7148 1.9572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7148 1.9572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7148 2.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7148 2.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1598 2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1598 2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2697 2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2697 2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9115 2.0995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9115 2.0995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6260 1.6870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6260 1.6870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5826 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5826 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3795 -0.5791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3795 -0.5791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 19 3 1 0 0 0 0 | + | 19 3 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 36 41 1 0 0 0 0 | + | 36 41 1 0 0 0 0 |
| − | 37 42 1 0 0 0 0 | + | 37 42 1 0 0 0 0 |
| − | 27 34 1 0 0 0 0 | + | 27 34 1 0 0 0 0 |
| − | 6 43 1 0 0 0 0 | + | 6 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 15 48 1 0 0 0 0 | + | 15 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 38 50 1 0 0 0 0 | + | 38 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 48 49 | + | M SAL 1 2 48 49 |
| − | M SBL 1 1 52 | + | M SBL 1 1 52 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 52 -5.5984 4.1350 | + | M SBV 1 52 -5.5984 4.1350 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 50 51 | + | M SAL 2 2 50 51 |
| − | M SBL 2 1 54 | + | M SBL 2 1 54 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 54 -5.7827 3.8945 | + | M SBV 2 54 -5.7827 3.8945 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FABGI0017 | + | ID FL5FABGI0017 |
| − | KNApSAcK_ID C00006054 | + | KNApSAcK_ID C00006054 |
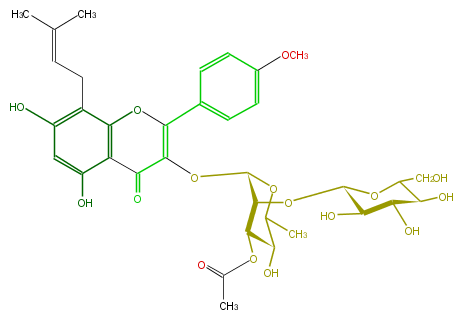
| − | NAME Sagittatoside C | + | NAME Sagittatoside C |
| − | CAS_RN 118525-37-4 | + | CAS_RN 118525-37-4 |
| − | FORMULA C35H42O16 | + | FORMULA C35H42O16 |
| − | EXACTMASS 718.247285296 | + | EXACTMASS 718.247285296 |
| − | AVERAGEMASS 718.69838 | + | AVERAGEMASS 718.69838 |
| − | SMILES C(C(O)1)(C(O)C(OC(C2OC(=C4c(c5)ccc(OC)c5)C(=O)c(c(O)3)c(O4)c(CC=C(C)C)c(c3)O)C(OC(C)=O)C(O)C(C)O2)OC(CO)1)O | + | SMILES C(C(O)1)(C(O)C(OC(C2OC(=C4c(c5)ccc(OC)c5)C(=O)c(c(O)3)c(O4)c(CC=C(C)C)c(c3)O)C(OC(C)=O)C(O)C(C)O2)OC(CO)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
51 55 0 0 0 0 0 0 0 0999 V2000
-3.7150 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7150 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1587 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6024 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6024 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1587 0.9940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0461 -0.2908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4898 0.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4898 0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0461 0.9940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0461 -0.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7893 1.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2223 0.7903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3447 1.1176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3447 1.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2223 2.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7893 1.7723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3795 1.0564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1587 -0.8535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9058 -0.3585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1469 -1.4622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6823 -1.7713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5124 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6823 -0.5788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1469 -0.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3167 -0.8641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9733 -0.8641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2844 -1.9755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5531 -2.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0410 -1.4821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3391 -2.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7516 -2.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3391 -2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0673 -0.5587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4690 -1.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0475 -0.8640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6057 -0.8579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2001 -0.4523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6940 -0.7194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8428 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3789 -1.4204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0612 -0.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1587 1.6361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7148 1.9572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7148 2.5979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1598 2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2697 2.9183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9115 2.0995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6260 1.6870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5826 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3795 -0.5791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
4 3 1 0 0 0 0
1 18 1 0 0 0 0
19 3 1 0 0 0 0
8 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
25 20 1 0 0 0 0
28 31 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
35 40 1 0 0 0 0
36 41 1 0 0 0 0
37 42 1 0 0 0 0
27 34 1 0 0 0 0
6 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
45 47 1 0 0 0 0
15 48 1 0 0 0 0
48 49 1 0 0 0 0
38 50 1 0 0 0 0
50 51 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 48 49
M SBL 1 1 52
M SMT 1 OCH3
M SBV 1 52 -5.5984 4.1350
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 50 51
M SBL 2 1 54
M SMT 2 CH2OH
M SBV 2 54 -5.7827 3.8945
S SKP 8
ID FL5FABGI0017
KNApSAcK_ID C00006054
NAME Sagittatoside C
CAS_RN 118525-37-4
FORMULA C35H42O16
EXACTMASS 718.247285296
AVERAGEMASS 718.69838
SMILES C(C(O)1)(C(O)C(OC(C2OC(=C4c(c5)ccc(OC)c5)C(=O)c(c(O)3)c(O4)c(CC=C(C)C)c(c3)O)C(OC(C)=O)C(O)C(C)O2)OC(CO)1)O
M END