Mol:FL5FACGL0020
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3008 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3008 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3008 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3008 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7445 0.4241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7445 0.4241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1882 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1882 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1882 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1882 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7445 1.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7445 1.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6319 0.4241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6319 0.4241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0756 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0756 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0756 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0756 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6319 1.7088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6319 1.7088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6319 -0.0767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6319 -0.0767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3752 1.8325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3752 1.8325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1918 1.5051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1918 1.5051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7588 1.8325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7588 1.8325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7588 2.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7588 2.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1918 2.8145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1918 2.8145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3752 2.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3752 2.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7445 -0.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7445 -0.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3656 2.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3656 2.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5123 0.3358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5123 0.3358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1918 3.4690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1918 3.4690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9653 1.7713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9653 1.7713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6771 -0.6894 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.6771 -0.6894 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.0788 -1.2197 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0788 -1.2197 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.6572 -0.9947 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6572 -0.9947 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2154 -0.9887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2154 -0.9887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8098 -0.5830 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.8098 -0.5830 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.3037 -0.8501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3037 -0.8501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4526 -1.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4526 -1.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9887 -1.5511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9887 -1.5511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6728 -0.9487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6728 -0.9487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2313 -1.8505 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -1.2313 -1.8505 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.5919 -1.6855 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.5919 -1.6855 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.6224 -1.0323 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.6224 -1.0323 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.3032 -0.4686 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.3032 -0.4686 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.0044 -0.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0044 -0.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9426 -1.3967 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.9426 -1.3967 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -1.2313 -2.3698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2313 -2.3698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8504 -2.6500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8504 -2.6500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0366 -1.0243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0366 -1.0243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0474 -1.5162 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.0474 -1.5162 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.4458 -1.5162 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.4458 -1.5162 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.8782 -1.9345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8782 -1.9345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0260 -2.5212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.0260 -2.5212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.6277 -2.5213 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.6277 -2.5213 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.1953 -2.1029 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.1953 -2.1029 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.6384 -1.1072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6384 -1.1072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6270 -1.9016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6270 -1.9016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3895 -3.1691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3895 -3.1691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9166 -0.1550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9166 -0.1550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1988 0.6202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1988 0.6202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5545 -0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5545 -0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4458 -1.4520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4458 -1.4520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0046 -3.5210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0046 -3.5210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1281 -4.0024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1281 -4.0024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 1 22 1 0 0 0 0 | + | 1 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 24 29 1 0 0 0 0 | + | 24 29 1 0 0 0 0 |
| − | 25 30 1 0 0 0 0 | + | 25 30 1 0 0 0 0 |
| − | 26 31 1 0 0 0 0 | + | 26 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 20 1 0 0 0 0 | + | 35 20 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 46 48 1 0 0 0 0 | + | 46 48 1 0 0 0 0 |
| − | 45 49 1 0 0 0 0 | + | 45 49 1 0 0 0 0 |
| − | 42 40 1 0 0 0 0 | + | 42 40 1 0 0 0 0 |
| − | 23 47 1 0 0 0 0 | + | 23 47 1 0 0 0 0 |
| − | 27 50 1 0 0 0 0 | + | 27 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 37 52 1 0 0 0 0 | + | 37 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 44 54 1 0 0 0 0 | + | 44 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 54 55 | + | M SAL 3 2 54 55 |
| − | M SBL 3 1 59 | + | M SBL 3 1 59 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 59 1.3332 -3.0565 | + | M SVB 3 59 1.3332 -3.0565 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 57 -1.5545 -0.9987 | + | M SVB 2 57 -1.5545 -0.9987 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 55 3.2509 -0.5754 | + | M SVB 1 55 3.2509 -0.5754 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGL0020 | + | ID FL5FACGL0020 |
| − | KNApSAcK_ID C00005446 | + | KNApSAcK_ID C00005446 |
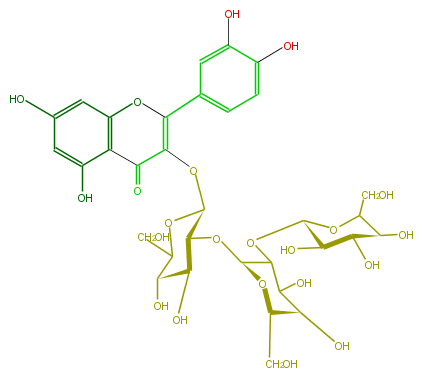
| − | NAME Quercetin 3-sophorotrioside | + | NAME Quercetin 3-sophorotrioside |
| − | CAS_RN 38681-85-5 | + | CAS_RN 38681-85-5 |
| − | FORMULA C33H40O22 | + | FORMULA C33H40O22 |
| − | EXACTMASS 788.201122964 | + | EXACTMASS 788.201122964 |
| − | AVERAGEMASS 788.6575 | + | AVERAGEMASS 788.6575 |
| − | SMILES [C@@H](O)([C@H]5CO)C(C(O[C@@H](O6)[C@H](O)[C@H](O)[C@H](O)C6CO)[C@@H](O5)O[C@@H]([C@@H](O)4)[C@@H](OC([C@@H]4O)CO)OC(=C(c(c3)ccc(O)c3O)1)C(=O)c(c2O)c(cc(O)c2)O1)O | + | SMILES [C@@H](O)([C@H]5CO)C(C(O[C@@H](O6)[C@H](O)[C@H](O)[C@H](O)C6CO)[C@@H](O5)O[C@@H]([C@@H](O)4)[C@@H](OC([C@@H]4O)CO)OC(=C(c(c3)ccc(O)c3O)1)C(=O)c(c2O)c(cc(O)c2)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-3.3008 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3008 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7445 0.4241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1882 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1882 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7445 1.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6319 0.4241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0756 0.7453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0756 1.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6319 1.7088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6319 -0.0767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3752 1.8325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1918 1.5051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7588 1.8325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7588 2.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1918 2.8145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3752 2.4872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7445 -0.2180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3656 2.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5123 0.3358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1918 3.4690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9653 1.7713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6771 -0.6894 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.0788 -1.2197 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.6572 -0.9947 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2154 -0.9887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8098 -0.5830 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.3037 -0.8501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4526 -1.2502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9887 -1.5511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6728 -0.9487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2313 -1.8505 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.5919 -1.6855 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6224 -1.0323 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.3032 -0.4686 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.0044 -0.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9426 -1.3967 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-1.2313 -2.3698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8504 -2.6500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0366 -1.0243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0474 -1.5162 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.4458 -1.5162 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.8782 -1.9345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0260 -2.5212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6277 -2.5213 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.1953 -2.1029 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.6384 -1.1072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6270 -1.9016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3895 -3.1691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9166 -0.1550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1988 0.6202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5545 -0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4458 -1.4520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0046 -3.5210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1281 -4.0024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
16 21 1 0 0 0 0
1 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
24 29 1 0 0 0 0
25 30 1 0 0 0 0
26 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 20 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
46 48 1 0 0 0 0
45 49 1 0 0 0 0
42 40 1 0 0 0 0
23 47 1 0 0 0 0
27 50 1 0 0 0 0
50 51 1 0 0 0 0
37 52 1 0 0 0 0
52 53 1 0 0 0 0
44 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 54 55
M SBL 3 1 59
M SMT 3 CH2OH
M SVB 3 59 1.3332 -3.0565
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 57
M SMT 2 CH2OH
M SVB 2 57 -1.5545 -0.9987
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 55
M SMT 1 CH2OH
M SVB 1 55 3.2509 -0.5754
S SKP 8
ID FL5FACGL0020
KNApSAcK_ID C00005446
NAME Quercetin 3-sophorotrioside
CAS_RN 38681-85-5
FORMULA C33H40O22
EXACTMASS 788.201122964
AVERAGEMASS 788.6575
SMILES [C@@H](O)([C@H]5CO)C(C(O[C@@H](O6)[C@H](O)[C@H](O)[C@H](O)C6CO)[C@@H](O5)O[C@@H]([C@@H](O)4)[C@@H](OC([C@@H]4O)CO)OC(=C(c(c3)ccc(O)c3O)1)C(=O)c(c2O)c(cc(O)c2)O1)O
M END