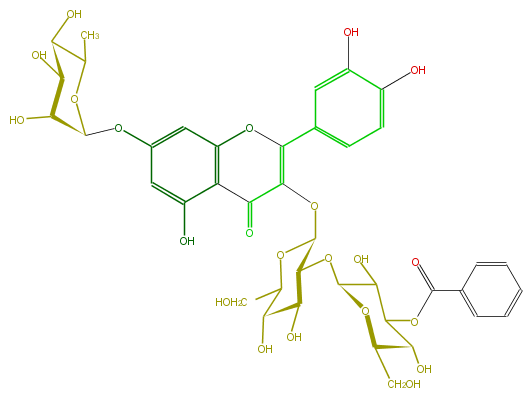
Mol:FL5FACGL0076
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 62 68 0 0 0 0 0 0 0 0999 V2000 | + | 62 68 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.6089 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6089 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6089 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6089 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8944 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8944 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1799 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1799 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1799 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1799 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8944 1.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8944 1.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4653 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4653 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2492 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2492 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2492 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2492 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4653 1.8161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4653 1.8161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4653 -0.4773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4653 -0.4773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9635 1.8160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9635 1.8160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6918 1.3956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6918 1.3956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4201 1.8160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4201 1.8160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4201 2.6569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4201 2.6569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6918 3.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6918 3.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9635 2.6569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9635 2.6569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3232 1.8160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3232 1.8160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1481 3.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1481 3.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9632 0.1189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9632 0.1189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8944 -0.6588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8944 -0.6588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6918 3.9180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6918 3.9180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1548 -2.3233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1548 -2.3233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6329 -2.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6329 -2.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5986 -1.3459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5986 -1.3459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9706 -0.6233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9706 -0.6233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2137 -0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2137 -0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2228 -1.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2228 -1.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1946 -3.0144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1946 -3.0144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4280 -2.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4280 -2.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2657 -1.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2657 -1.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3032 -1.6170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3032 -1.6170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4827 -1.6170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4827 -1.6170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0724 -2.1876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0724 -2.1876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2741 -2.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2741 -2.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0947 -2.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0947 -2.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5049 -2.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5049 -2.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1547 -2.4172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1547 -2.4172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2793 -3.4581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2793 -3.4581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9031 -1.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9031 -1.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7952 2.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7952 2.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0657 1.6443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0657 1.6443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2973 2.4542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2973 2.4542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0657 3.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0657 3.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7952 3.6902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7952 3.6902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5638 2.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5638 2.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3804 4.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3804 4.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0770 3.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0770 3.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1354 3.4174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1354 3.4174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5422 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5422 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1728 -1.1062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1728 -1.1062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1643 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1643 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4910 -2.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4910 -2.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1447 -2.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1447 -2.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4714 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4714 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1447 -1.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1447 -1.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4910 -1.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4910 -1.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4714 2.0009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4714 2.0009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4531 -1.9842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4531 -1.9842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6095 -2.5433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6095 -2.5433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6468 -3.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6468 -3.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5646 -4.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5646 -4.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 20 1 0 0 0 0 | + | 26 20 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 33 31 1 0 0 0 0 | + | 33 31 1 0 0 0 0 |
| − | 40 32 1 0 0 0 0 | + | 40 32 1 0 0 0 0 |
| − | 42 41 1 1 0 0 0 | + | 42 41 1 1 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 46 41 1 1 0 0 0 | + | 46 41 1 1 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 44 48 1 0 0 0 0 | + | 44 48 1 0 0 0 0 |
| − | 46 49 1 0 0 0 0 | + | 46 49 1 0 0 0 0 |
| − | 18 42 1 0 0 0 0 | + | 18 42 1 0 0 0 0 |
| − | 38 50 1 0 0 0 0 | + | 38 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 50 52 1 0 0 0 0 | + | 50 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 55 2 0 0 0 0 | + | 54 55 2 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 56 57 2 0 0 0 0 | + | 56 57 2 0 0 0 0 |
| − | 57 52 1 0 0 0 0 | + | 57 52 1 0 0 0 0 |
| − | 41 58 1 0 0 0 0 | + | 41 58 1 0 0 0 0 |
| − | 59 60 1 0 0 0 0 | + | 59 60 1 0 0 0 0 |
| − | 28 59 1 0 0 0 0 | + | 28 59 1 0 0 0 0 |
| − | 61 62 1 0 0 0 0 | + | 61 62 1 0 0 0 0 |
| − | 35 61 1 0 0 0 0 | + | 35 61 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 59 60 | + | M SAL 1 2 59 60 |
| − | M SBL 1 1 66 | + | M SBL 1 1 66 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 66 0.6759 0.2754 | + | M SBV 1 66 0.6759 0.2754 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 61 62 | + | M SAL 2 2 61 62 |
| − | M SBL 2 1 68 | + | M SBL 2 1 68 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 68 -0.3728 0.8030 | + | M SBV 2 68 -0.3728 0.8030 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGL0076 | + | ID FL5FACGL0076 |
| − | FORMULA C40H44O22 | + | FORMULA C40H44O22 |
| − | EXACTMASS 876.232423092 | + | EXACTMASS 876.232423092 |
| − | AVERAGEMASS 876.7641600000001 | + | AVERAGEMASS 876.7641600000001 |
| − | SMILES C(OC(C(O)6)OC(C(O)C6OC(c(c7)cccc7)=O)CO)(C1OC(C4=O)=C(c(c5)ccc(c5O)O)Oc(c24)cc(OC(C(O)3)OC(C)C(O)C3O)cc2O)C(O)C(O)C(O1)CO | + | SMILES C(OC(C(O)6)OC(C(O)C6OC(c(c7)cccc7)=O)CO)(C1OC(C4=O)=C(c(c5)ccc(c5O)O)Oc(c24)cc(OC(C(O)3)OC(C)C(O)C3O)cc2O)C(O)C(O)C(O1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
62 68 0 0 0 0 0 0 0 0999 V2000
-2.6089 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6089 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8944 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1799 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1799 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8944 1.8161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4653 0.1660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2492 0.5786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2492 1.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4653 1.8161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4653 -0.4773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9635 1.8160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6918 1.3956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4201 1.8160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4201 2.6569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6918 3.0773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9635 2.6569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3232 1.8160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1481 3.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9632 0.1189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8944 -0.6588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6918 3.9180 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1548 -2.3233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6329 -2.0464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5986 -1.3459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9706 -0.6233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2137 -0.9555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2228 -1.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1946 -3.0144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4280 -2.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2657 -1.0299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3032 -1.6170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4827 -1.6170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0724 -2.1876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2741 -2.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0947 -2.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5049 -2.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1547 -2.4172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2793 -3.4581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9031 -1.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7952 2.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0657 1.6443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2973 2.4542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0657 3.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7952 3.6902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5638 2.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3804 4.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0770 3.8498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1354 3.4174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5422 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1728 -1.1062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1643 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4910 -2.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1447 -2.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4714 -1.7460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1447 -1.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4910 -1.1800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4714 2.0009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4531 -1.9842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6095 -2.5433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6468 -3.7908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5646 -4.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 20 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
36 39 1 0 0 0 0
33 31 1 0 0 0 0
40 32 1 0 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 1 0 0 0
46 41 1 1 0 0 0
45 47 1 0 0 0 0
44 48 1 0 0 0 0
46 49 1 0 0 0 0
18 42 1 0 0 0 0
38 50 1 0 0 0 0
50 51 2 0 0 0 0
50 52 1 0 0 0 0
52 53 2 0 0 0 0
53 54 1 0 0 0 0
54 55 2 0 0 0 0
55 56 1 0 0 0 0
56 57 2 0 0 0 0
57 52 1 0 0 0 0
41 58 1 0 0 0 0
59 60 1 0 0 0 0
28 59 1 0 0 0 0
61 62 1 0 0 0 0
35 61 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 59 60
M SBL 1 1 66
M SMT 1 ^ CH2OH
M SBV 1 66 0.6759 0.2754
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 61 62
M SBL 2 1 68
M SMT 2 CH2OH
M SBV 2 68 -0.3728 0.8030
S SKP 5
ID FL5FACGL0076
FORMULA C40H44O22
EXACTMASS 876.232423092
AVERAGEMASS 876.7641600000001
SMILES C(OC(C(O)6)OC(C(O)C6OC(c(c7)cccc7)=O)CO)(C1OC(C4=O)=C(c(c5)ccc(c5O)O)Oc(c24)cc(OC(C(O)3)OC(C)C(O)C3O)cc2O)C(O)C(O)C(O1)CO
M END