Mol:FL5FACGS0042
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.9514 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9514 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9514 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9514 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3951 0.4980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3951 0.4980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8388 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8388 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8388 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8388 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3951 1.7827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3951 1.7827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2825 0.4980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2825 0.4980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7262 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7262 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7262 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7262 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2825 1.7827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2825 1.7827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2825 -0.0028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2825 -0.0028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0257 1.9064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0257 1.9064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5412 1.5790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5412 1.5790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1082 1.9064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1082 1.9064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1082 2.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1082 2.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5412 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5412 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0257 2.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0257 2.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3951 -0.1441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3951 -0.1441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7150 2.9114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7150 2.9114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1629 0.4097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1629 0.4097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5412 3.5429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5412 3.5429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6159 1.8452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6159 1.8452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6217 -0.8285 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.6217 -0.8285 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.2499 -1.1912 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2499 -1.1912 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.0505 -0.4937 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.0505 -0.4937 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.2499 0.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2499 0.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6217 0.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.6217 0.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.8209 -0.1267 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.8209 -0.1267 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.5913 -0.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5913 -0.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7830 -1.4307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7830 -1.4307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0983 -1.7569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0983 -1.7569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6708 -0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6708 -0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2193 -1.5708 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.2193 -1.5708 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.9565 -2.0260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.9565 -2.0260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4619 -1.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4619 -1.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9703 -2.0260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.9703 -2.0260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2331 -1.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2331 -1.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7277 -1.7152 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.7277 -1.7152 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.2634 -1.0675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2634 -1.0675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7640 -1.3006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7640 -1.3006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1750 -1.2350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1750 -1.2350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9703 -2.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9703 -2.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9108 -2.7745 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.9108 -2.7745 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.2690 -3.2473 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2690 -3.2473 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7848 -3.0467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7848 -3.0467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2825 -3.0414 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2825 -3.0414 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9209 -2.6796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9209 -2.6796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3940 -2.8688 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3940 -2.8688 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.4764 -2.9326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4764 -2.9326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1507 -3.7936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1507 -3.7936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0804 -3.5429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0804 -3.5429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6159 -2.4640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6159 -2.4640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4567 -2.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4567 -2.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6862 -3.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6862 -3.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 1 22 1 0 0 0 0 | + | 1 22 1 0 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 23 1 1 0 0 0 | + | 28 23 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 27 20 1 0 0 0 0 | + | 27 20 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 52 42 1 0 0 0 0 | + | 52 42 1 0 0 0 0 |
| − | 34 31 1 0 0 0 0 | + | 34 31 1 0 0 0 0 |
| − | 48 53 1 0 0 0 0 | + | 48 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 58 2.1217 -2.1368 | + | M SVB 1 58 2.1217 -2.1368 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0042 | + | ID FL5FACGS0042 |
| − | KNApSAcK_ID C00005458 | + | KNApSAcK_ID C00005458 |
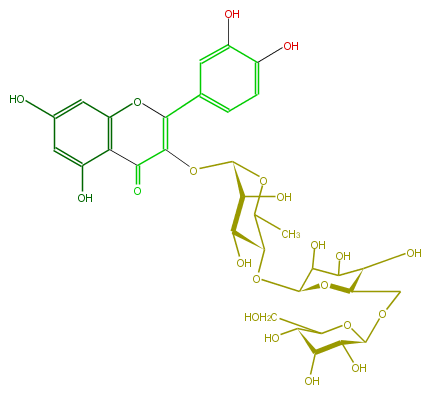
| − | NAME Quercetin 3-glucosyl-(1->6)-glucosyl-(1->4)-rhamnoside | + | NAME Quercetin 3-glucosyl-(1->6)-glucosyl-(1->4)-rhamnoside |
| − | CAS_RN 59262-55-4 | + | CAS_RN 59262-55-4 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES OC([C@H]5O)C([C@@H](O[C@@H]5CO[C@H](O6)[C@H]([C@@H](O)[C@H](C(CO)6)O)O)O[C@H](C(C)4)[C@@H](O)[C@H](O)[C@H](O4)OC(C(=O)3)=C(Oc(c23)cc(cc(O)2)O)c(c1)ccc(O)c1O)O | + | SMILES OC([C@H]5O)C([C@@H](O[C@@H]5CO[C@H](O6)[C@H]([C@@H](O)[C@H](C(CO)6)O)O)O[C@H](C(C)4)[C@@H](O)[C@H](O)[C@H](O4)OC(C(=O)3)=C(Oc(c23)cc(cc(O)2)O)c(c1)ccc(O)c1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.9514 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9514 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3951 0.4980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8388 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8388 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3951 1.7827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2825 0.4980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7262 0.8192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7262 1.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2825 1.7827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2825 -0.0028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0257 1.9064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5412 1.5790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1082 1.9064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1082 2.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5412 2.8884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0257 2.5610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3951 -0.1441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7150 2.9114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1629 0.4097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5412 3.5429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6159 1.8452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6217 -0.8285 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.2499 -1.1912 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.0505 -0.4937 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.2499 0.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6217 0.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.8209 -0.1267 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.5913 -0.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7830 -1.4307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0983 -1.7569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6708 -0.8518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2193 -1.5708 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9565 -2.0260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4619 -1.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9703 -2.0260 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2331 -1.5708 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7277 -1.7152 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.2634 -1.0675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7640 -1.3006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1750 -1.2350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9703 -2.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9108 -2.7745 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.2690 -3.2473 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7848 -3.0467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2825 -3.0414 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9209 -2.6796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3940 -2.8688 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.4764 -2.9326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1507 -3.7936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0804 -3.5429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6159 -2.4640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4567 -2.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6862 -3.1577 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
16 21 1 0 0 0 0
1 22 1 0 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 1 0 0 0
28 23 1 1 0 0 0
28 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
27 20 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
38 40 1 0 0 0 0
37 41 1 0 0 0 0
36 42 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 52 1 0 0 0 0
52 42 1 0 0 0 0
34 31 1 0 0 0 0
48 53 1 0 0 0 0
53 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 58
M SMT 1 CH2OH
M SVB 1 58 2.1217 -2.1368
S SKP 8
ID FL5FACGS0042
KNApSAcK_ID C00005458
NAME Quercetin 3-glucosyl-(1->6)-glucosyl-(1->4)-rhamnoside
CAS_RN 59262-55-4
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES OC([C@H]5O)C([C@@H](O[C@@H]5CO[C@H](O6)[C@H]([C@@H](O)[C@H](C(CO)6)O)O)O[C@H](C(C)4)[C@@H](O)[C@H](O)[C@H](O4)OC(C(=O)3)=C(Oc(c23)cc(cc(O)2)O)c(c1)ccc(O)c1O)O
M END