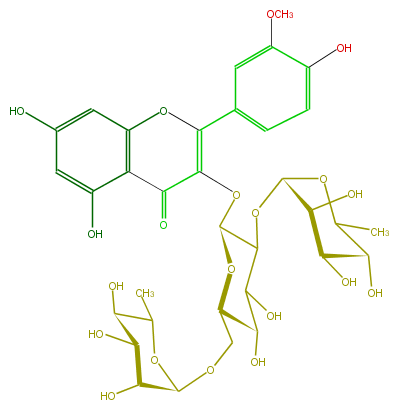
Mol:FL5FADGL0022
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.8250 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8250 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8250 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8250 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1105 -0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1105 -0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3961 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3961 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3961 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3961 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1105 1.3110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1105 1.3110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6817 -0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6817 -0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0327 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0327 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0327 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0327 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6817 1.3110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6817 1.3110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6817 -0.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6817 -0.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7469 1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7469 1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3720 0.8904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3720 0.8904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2032 1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2032 1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2032 2.1516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2032 2.1516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4751 2.5720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4751 2.5720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7469 2.1516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7469 2.1516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5391 1.3108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5391 1.3108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7879 -0.3860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7879 -0.3860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1105 -1.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1105 -1.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8630 2.5654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8630 2.5654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1850 -1.4694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1850 -1.4694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4365 -1.0374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4365 -1.0374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6740 -1.8683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6740 -1.8683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4365 -2.7044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4365 -2.7044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1850 -3.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1850 -3.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9475 -2.3055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9475 -2.3055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1583 -0.7315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1583 -0.7315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4591 -2.7935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4591 -2.7935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1451 -3.7226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1451 -3.7226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6930 -3.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6930 -3.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1641 -4.1845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1641 -4.1845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3963 -4.3200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3963 -4.3200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8540 -3.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8540 -3.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9106 -2.9067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9106 -2.9067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6786 -2.7711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6786 -2.7711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2208 -3.4025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2208 -3.4025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7087 -2.2023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7087 -2.2023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1695 -2.3312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1695 -2.3312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9491 -3.1492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9491 -3.1492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7290 -4.4010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7290 -4.4010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2653 -3.8860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2653 -3.8860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4797 -1.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4797 -1.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4113 -1.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4113 -1.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7417 -0.9685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7417 -0.9685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5129 -0.0602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5129 -0.0602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6843 -0.0600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6843 -0.0600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2508 -0.7078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2508 -0.7078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0882 -0.3590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0882 -0.3590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9647 -2.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9647 -2.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4837 -2.3442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4837 -2.3442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5391 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5391 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4692 3.2486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4692 3.2486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0360 4.4010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0360 4.4010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 32 1 1 0 0 0 | + | 37 32 1 1 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | 33 42 1 0 0 0 0 | + | 33 42 1 0 0 0 0 |
| − | 42 31 1 0 0 0 0 | + | 42 31 1 0 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 1 0 0 0 | + | 47 48 1 1 0 0 0 |
| − | 48 43 1 1 0 0 0 | + | 48 43 1 1 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 43 50 1 0 0 0 0 | + | 43 50 1 0 0 0 0 |
| − | 44 51 1 0 0 0 0 | + | 44 51 1 0 0 0 0 |
| − | 45 52 1 0 0 0 0 | + | 45 52 1 0 0 0 0 |
| − | 47 28 1 0 0 0 0 | + | 47 28 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 16 53 1 0 0 0 0 | + | 16 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 59 0.0059 -0.6766 | + | M SBV 1 59 0.0059 -0.6766 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0022 | + | ID FL5FADGL0022 |
| − | FORMULA C34H42O20 | + | FORMULA C34H42O20 |
| − | EXACTMASS 770.226943784 | + | EXACTMASS 770.226943784 |
| − | AVERAGEMASS 770.6852799999999 | + | AVERAGEMASS 770.6852799999999 |
| − | SMILES C(C(O)1)(C(O)C(C)OC1OC(C3OC(=C5c(c6)cc(OC)c(O)c6)C(=O)c(c(O5)4)c(cc(c4)O)O)C(C(C(O3)COC(O2)C(O)C(C(C2C)O)O)O)O)O | + | SMILES C(C(O)1)(C(O)C(C)OC1OC(C3OC(=C5c(c6)cc(OC)c(O)c6)C(=O)c(c(O5)4)c(cc(c4)O)O)C(C(C(O3)COC(O2)C(O)C(C(C2C)O)O)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.8250 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8250 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1105 -0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3961 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3961 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1105 1.3110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6817 -0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0327 0.0735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0327 0.8985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6817 1.3110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6817 -0.9821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7469 1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3720 0.8904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2032 1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2032 2.1516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4751 2.5720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7469 2.1516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5391 1.3108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7879 -0.3860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1105 -1.1636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8630 2.5654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1850 -1.4694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4365 -1.0374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6740 -1.8683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4365 -2.7044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1850 -3.1365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9475 -2.3055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1583 -0.7315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4591 -2.7935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1451 -3.7226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6930 -3.3428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1641 -4.1845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3963 -4.3200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8540 -3.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9106 -2.9067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6786 -2.7711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2208 -3.4025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7087 -2.2023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1695 -2.3312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9491 -3.1492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7290 -4.4010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2653 -3.8860 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4797 -1.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4113 -1.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7417 -0.9685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5129 -0.0602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6843 -0.0600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2508 -0.7078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0882 -0.3590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9647 -2.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4837 -2.3442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5391 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4692 3.2486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0360 4.4010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
23 19 1 0 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 32 1 1 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
37 40 1 0 0 0 0
32 41 1 0 0 0 0
33 42 1 0 0 0 0
42 31 1 0 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 48 1 1 0 0 0
48 43 1 1 0 0 0
48 49 1 0 0 0 0
43 50 1 0 0 0 0
44 51 1 0 0 0 0
45 52 1 0 0 0 0
47 28 1 0 0 0 0
53 54 1 0 0 0 0
16 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 59
M SMT 1 OCH3
M SBV 1 59 0.0059 -0.6766
S SKP 5
ID FL5FADGL0022
FORMULA C34H42O20
EXACTMASS 770.226943784
AVERAGEMASS 770.6852799999999
SMILES C(C(O)1)(C(O)C(C)OC1OC(C3OC(=C5c(c6)cc(OC)c(O)c6)C(=O)c(c(O5)4)c(cc(c4)O)O)C(C(C(O3)COC(O2)C(O)C(C(C2C)O)O)O)O)O
M END