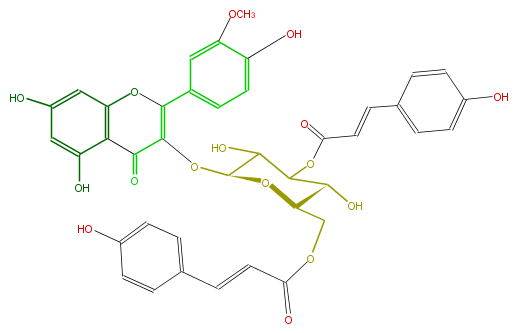
Mol:FL5FADGL0039
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.8235 -0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8235 -0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5521 -1.3299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5521 -1.3299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9122 -1.3859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9122 -1.3859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5437 -0.8597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5437 -0.8597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8152 -0.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8152 -0.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4551 -0.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4551 -0.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9038 -0.9157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9038 -0.9157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5354 -0.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5354 -0.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8068 0.1926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8068 0.1926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4468 0.2486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4468 0.2486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6921 -1.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6921 -1.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4385 0.7186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4385 0.7186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7863 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7863 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4109 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4109 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6876 1.7912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6876 1.7912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3397 1.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3397 1.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7152 1.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7152 1.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7154 -0.6144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7154 -0.6144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3444 0.1719 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.3444 0.1719 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.0432 -0.4995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.0432 -0.4995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.7023 -0.2865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7023 -0.2865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4522 -0.4995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.4522 -0.4995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.8400 0.1719 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.8400 0.1719 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.0944 -0.0411 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.0944 -0.0411 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.3826 -0.0229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3826 -0.0229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3633 0.4245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3633 0.4245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4542 0.0073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4542 0.0073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1140 2.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1140 2.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6408 -1.9677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6408 -1.9677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4611 -0.8261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4611 -0.8261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0812 -0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0812 -0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1594 -1.2925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1594 -1.2925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8913 -1.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8913 -1.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2737 -2.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2737 -2.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1051 -1.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1051 -1.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7126 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7126 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0726 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0726 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4127 -2.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4127 -2.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0930 -2.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0930 -2.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4331 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4331 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0930 -3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0930 -3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4127 -3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4127 -3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1124 -2.6324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1124 -2.6324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3633 0.9770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3633 0.9770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8960 1.1022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8960 1.1022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9405 1.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9405 1.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9405 1.9251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9405 1.9251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4402 2.2136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4402 2.2136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0222 1.8776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0222 1.8776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6041 2.2136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6041 2.2136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6041 2.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6041 2.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0222 3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0222 3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4402 2.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4402 2.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1852 3.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1852 3.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3263 2.4902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3263 2.4902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3056 3.4900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3056 3.4900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 26 44 1 0 0 0 0 | + | 26 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 48 1 0 0 0 0 | + | 53 48 1 0 0 0 0 |
| − | 51 54 1 0 0 0 0 | + | 51 54 1 0 0 0 0 |
| − | 16 55 1 0 0 0 0 | + | 16 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 55 56 | + | M SAL 1 2 55 56 |
| − | M SBL 1 1 60 | + | M SBL 1 1 60 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 60 0.0574 2.7524 | + | M SVB 1 60 0.0574 2.7524 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGL0039 | + | ID FL5FADGL0039 |
| − | KNApSAcK_ID C00006010 | + | KNApSAcK_ID C00006010 |
| − | NAME Isorhamnetin 3-(3'',6''-di-p-coumarylglucoside) | + | NAME Isorhamnetin 3-(3'',6''-di-p-coumarylglucoside) |
| − | CAS_RN 104582-31-2 | + | CAS_RN 104582-31-2 |
| − | FORMULA C40H34O16 | + | FORMULA C40H34O16 |
| − | EXACTMASS 770.18468504 | + | EXACTMASS 770.18468504 |
| − | AVERAGEMASS 770.68836 | + | AVERAGEMASS 770.68836 |
| − | SMILES c(O)(c1)c(C3=O)c(OC(=C3O[C@@H](C(O)4)O[C@H](COC(=O)C=Cc(c6)ccc(O)c6)[C@H](O)C4OC(=O)C=Cc(c5)ccc(c5)O)c(c2)ccc(O)c2OC)cc(O)1 | + | SMILES c(O)(c1)c(C3=O)c(OC(=C3O[C@@H](C(O)4)O[C@H](COC(=O)C=Cc(c6)ccc(O)c6)[C@H](O)C4OC(=O)C=Cc(c5)ccc(c5)O)c(c2)ccc(O)c2OC)cc(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
-3.8235 -0.7478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5521 -1.3299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9122 -1.3859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5437 -0.8597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8152 -0.2776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4551 -0.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9038 -0.9157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5354 -0.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8068 0.1926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4468 0.2486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6921 -1.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4385 0.7186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7863 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4109 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6876 1.7912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3397 1.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7152 1.3120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7154 -0.6144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3444 0.1719 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.0432 -0.4995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.7023 -0.2865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4522 -0.4995 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.8400 0.1719 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.0944 -0.0411 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.3826 -0.0229 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3633 0.4245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4542 0.0073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1140 2.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6408 -1.9677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4611 -0.8261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0812 -0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1594 -1.2925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8913 -1.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2737 -2.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1051 -1.9525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7126 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0726 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4127 -2.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0930 -2.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4331 -2.6324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0930 -3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4127 -3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1124 -2.6324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3633 0.9770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8960 1.1022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9405 1.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9405 1.9251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4402 2.2136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0222 1.8776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6041 2.2136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6041 2.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0222 3.2216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4402 2.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1852 3.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3263 2.4902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3056 3.4900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 37 1 0 0 0 0
40 43 1 0 0 0 0
26 44 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 48 1 0 0 0 0
51 54 1 0 0 0 0
16 55 1 0 0 0 0
55 56 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 55 56
M SBL 1 1 60
M SMT 1 OCH3
M SVB 1 60 0.0574 2.7524
S SKP 8
ID FL5FADGL0039
KNApSAcK_ID C00006010
NAME Isorhamnetin 3-(3'',6''-di-p-coumarylglucoside)
CAS_RN 104582-31-2
FORMULA C40H34O16
EXACTMASS 770.18468504
AVERAGEMASS 770.68836
SMILES c(O)(c1)c(C3=O)c(OC(=C3O[C@@H](C(O)4)O[C@H](COC(=O)C=Cc(c6)ccc(O)c6)[C@H](O)C4OC(=O)C=Cc(c5)ccc(c5)O)c(c2)ccc(O)c2OC)cc(O)1
M END