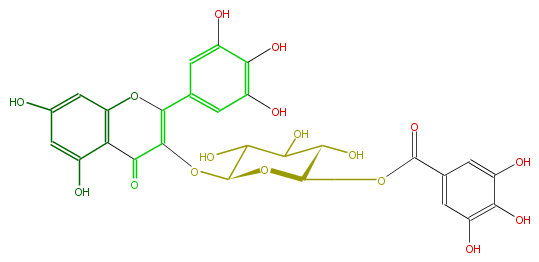
Mol:FL5FAGGL0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.3757 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3757 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3757 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3757 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8194 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8194 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2631 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2631 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2631 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2631 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8194 0.7157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8194 0.7157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7068 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7068 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1505 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1505 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1505 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1505 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7068 0.7157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7068 0.7157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7068 -1.0698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7068 -1.0698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5944 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5944 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0274 0.3883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0274 0.3883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4604 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4604 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4604 1.3703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4604 1.3703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0274 1.6976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0274 1.6976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5944 1.3703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5944 1.3703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5023 -0.7981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5023 -0.7981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4569 -0.3067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4569 -0.3067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8446 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8446 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0991 -0.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0991 -0.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6509 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6509 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0386 -0.3067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0386 -0.3067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2931 -0.5197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2931 -0.5197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1840 -0.5015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1840 -0.5015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5619 -0.0541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5619 -0.0541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6529 -0.4713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6529 -0.4713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1064 1.6976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1064 1.6976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8194 -1.2111 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8194 -1.2111 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9866 0.5931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9866 0.5931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2798 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2798 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2243 -0.9873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2243 -0.9873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8843 -0.5336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8843 -0.5336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8843 0.0855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8843 0.0855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0274 2.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0274 2.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4825 -0.9048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4825 -0.9048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4825 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4825 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9839 -1.7733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9839 -1.7733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4853 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4853 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4853 -0.9048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4853 -0.9048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9839 -0.6153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9839 -0.6153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9866 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9866 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9866 -1.7732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9866 -1.7732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9839 -2.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9839 -2.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1064 0.3884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1064 0.3884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 16 35 1 0 0 0 0 | + | 16 35 1 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 38 44 1 0 0 0 0 | + | 38 44 1 0 0 0 0 |
| − | 14 45 1 0 0 0 0 | + | 14 45 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAGGL0010 | + | ID FL5FAGGL0010 |
| − | KNApSAcK_ID C00006040 | + | KNApSAcK_ID C00006040 |
| − | NAME Myricetin 3-(6''-galloylglucoside) | + | NAME Myricetin 3-(6''-galloylglucoside) |
| − | CAS_RN 56317-07-8 | + | CAS_RN 56317-07-8 |
| − | FORMULA C28H24O17 | + | FORMULA C28H24O17 |
| − | EXACTMASS 632.101349342 | + | EXACTMASS 632.101349342 |
| − | AVERAGEMASS 632.47996 | + | AVERAGEMASS 632.47996 |
| − | SMILES C(OC(=O)c(c5)cc(O)c(c(O)5)O)C(O1)C(O)C(C(C1OC(C3=O)=C(c(c4)cc(c(O)c4O)O)Oc(c23)cc(cc(O)2)O)O)O | + | SMILES C(OC(=O)c(c5)cc(O)c(c(O)5)O)C(O1)C(O)C(C(C1OC(C3=O)=C(c(c4)cc(c(O)c4O)O)Oc(c23)cc(cc(O)2)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-4.3757 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3757 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8194 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2631 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2631 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8194 0.7157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7068 -0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1505 -0.2478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1505 0.3946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7068 0.7157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7068 -1.0698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5944 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0274 0.3883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4604 0.7156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4604 1.3703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0274 1.6976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5944 1.3703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5023 -0.7981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4569 -0.3067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8446 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0991 -0.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6509 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0386 -0.3067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2931 -0.5197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1840 -0.5015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5619 -0.0541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6529 -0.4713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1064 1.6976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8194 -1.2111 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9866 0.5931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2798 -0.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2243 -0.9873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8843 -0.5336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8843 0.0855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0274 2.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4825 -0.9048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4825 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9839 -1.7733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4853 -1.4838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4853 -0.9048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9839 -0.6153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9866 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9866 -1.7732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9839 -2.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1064 0.3884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
16 35 1 0 0 0 0
33 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
40 42 1 0 0 0 0
39 43 1 0 0 0 0
38 44 1 0 0 0 0
14 45 1 0 0 0 0
S SKP 8
ID FL5FAGGL0010
KNApSAcK_ID C00006040
NAME Myricetin 3-(6''-galloylglucoside)
CAS_RN 56317-07-8
FORMULA C28H24O17
EXACTMASS 632.101349342
AVERAGEMASS 632.47996
SMILES C(OC(=O)c(c5)cc(O)c(c(O)5)O)C(O1)C(O)C(C(C1OC(C3=O)=C(c(c4)cc(c(O)c4O)O)Oc(c23)cc(cc(O)2)O)O)O
M END