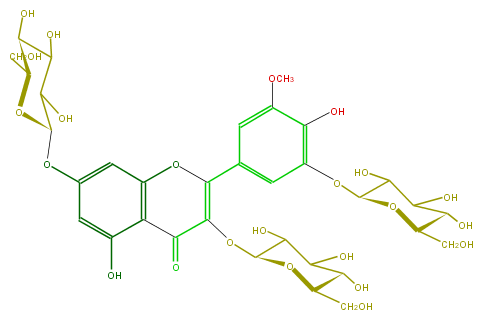
Mol:FL5FAHGL0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 57 62 0 0 0 0 0 0 0 0999 V2000 | + | 57 62 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.0253 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0253 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0253 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0253 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3106 -1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3106 -1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5959 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5959 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5959 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5959 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3106 0.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3106 0.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8812 -1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8812 -1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1665 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1665 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1665 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1665 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8812 0.4165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8812 0.4165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8812 -1.8776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8812 -1.8776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5479 0.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5479 0.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2763 -0.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2763 -0.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0047 0.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0047 0.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0047 1.2575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0047 1.2575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2763 1.6780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2763 1.6780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5479 1.2575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5479 1.2575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7398 0.4164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7398 0.4164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6649 1.6098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6649 1.6098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3398 -1.3210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3398 -1.3210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3106 -2.0590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3106 -2.0590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7329 -0.0041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7329 -0.0041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8967 0.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8967 0.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2273 -0.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2273 -0.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9773 -0.5261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9773 -0.5261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5185 -1.0839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5185 -1.0839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1880 -0.6975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1880 -0.6975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4381 -0.5098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4381 -0.5098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3454 0.2389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3454 0.2389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1825 -0.3191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1825 -0.3191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5021 -0.9447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5021 -0.9447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5826 -1.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5826 -1.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9133 -1.6693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9133 -1.6693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6632 -1.8569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6632 -1.8569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2045 -2.4147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2045 -2.4147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8739 -2.0283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8739 -2.0283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1240 -1.8406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1240 -1.8406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0712 -1.0491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0712 -1.0491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8478 -1.6292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8478 -1.6292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2015 -2.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2015 -2.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3810 1.5823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3810 1.5823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6514 1.1611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6514 1.1611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8829 1.9711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8829 1.9711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6514 2.7862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6514 2.7862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3810 3.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3810 3.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1496 2.3974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1496 2.3974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2707 3.7985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2707 3.7985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6897 3.3158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6897 3.3158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4069 1.4537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4069 1.4537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6250 2.8414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6250 2.8414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5780 1.9799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5780 1.9799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1302 -1.3721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1302 -1.3721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5780 -2.3326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5780 -2.3326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2719 2.3329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2719 2.3329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8390 3.4857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8390 3.4857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8716 -2.7386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8716 -2.7386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8716 -3.7985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8716 -3.7985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 24 22 1 0 0 0 0 | + | 24 22 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 42 41 1 1 0 0 0 | + | 42 41 1 1 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 46 41 1 1 0 0 0 | + | 46 41 1 1 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 44 48 1 0 0 0 0 | + | 44 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 18 42 1 0 0 0 0 | + | 18 42 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 46 50 1 0 0 0 0 | + | 46 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 26 52 1 0 0 0 0 | + | 26 52 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 16 54 1 0 0 0 0 | + | 16 54 1 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | 35 56 1 0 0 0 0 | + | 35 56 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 56 0.4754 -0.4440 | + | M SBV 1 56 0.4754 -0.4440 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 58 -0.6116 0.2882 | + | M SBV 2 58 -0.6116 0.2882 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 54 55 | + | M SAL 3 2 54 55 |
| − | M SBL 3 1 60 | + | M SBL 3 1 60 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 60 0.0044 -0.6549 | + | M SBV 3 60 0.0044 -0.6549 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 56 57 | + | M SAL 4 2 56 57 |
| − | M SBL 4 1 62 | + | M SBL 4 1 62 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SBV 4 62 -0.6671 0.3239 | + | M SBV 4 62 -0.6671 0.3239 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAHGL0005 | + | ID FL5FAHGL0005 |
| − | FORMULA C34H42O23 | + | FORMULA C34H42O23 |
| − | EXACTMASS 818.2116876499999 | + | EXACTMASS 818.2116876499999 |
| − | AVERAGEMASS 818.6834799999999 | + | AVERAGEMASS 818.6834799999999 |
| − | SMILES O(C1CO)C(Oc(c6)cc(O)c(c63)C(C(=C(c(c4)cc(OC)c(c4OC(C(O)5)OC(C(C(O)5)O)CO)O)O3)OC(O2)C(C(C(O)C2CO)O)O)=O)C(O)C(O)C1O | + | SMILES O(C1CO)C(Oc(c6)cc(O)c(c63)C(C(=C(c(c4)cc(OC)c(c4OC(C(O)5)OC(C(C(O)5)O)CO)O)O3)OC(O2)C(C(C(O)C2CO)O)O)=O)C(O)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
57 62 0 0 0 0 0 0 0 0999 V2000
-3.0253 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0253 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3106 -1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5959 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5959 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3106 0.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8812 -1.2340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1665 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1665 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8812 0.4165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8812 -1.8776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5479 0.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2763 -0.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0047 0.4164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0047 1.2575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2763 1.6780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5479 1.2575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7398 0.4164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6649 1.6098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3398 -1.3210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3106 -2.0590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7329 -0.0041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8967 0.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2273 -0.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9773 -0.5261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5185 -1.0839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1880 -0.6975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4381 -0.5098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3454 0.2389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1825 -0.3191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5021 -0.9447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5826 -1.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9133 -1.6693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6632 -1.8569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2045 -2.4147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8739 -2.0283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1240 -1.8406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0712 -1.0491 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8478 -1.6292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2015 -2.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3810 1.5823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6514 1.1611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8829 1.9711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6514 2.7862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3810 3.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1496 2.3974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2707 3.7985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6897 3.3158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4069 1.4537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6250 2.8414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5780 1.9799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1302 -1.3721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5780 -2.3326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2719 2.3329 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8390 3.4857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8716 -2.7386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8716 -3.7985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
14 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
24 22 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
33 20 1 0 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 1 0 0 0
46 41 1 1 0 0 0
45 47 1 0 0 0 0
44 48 1 0 0 0 0
43 49 1 0 0 0 0
18 42 1 0 0 0 0
50 51 1 0 0 0 0
46 50 1 0 0 0 0
52 53 1 0 0 0 0
26 52 1 0 0 0 0
54 55 1 0 0 0 0
16 54 1 0 0 0 0
56 57 1 0 0 0 0
35 56 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 ^ CH2OH
M SBV 1 56 0.4754 -0.4440
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 CH2OH
M SBV 2 58 -0.6116 0.2882
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 54 55
M SBL 3 1 60
M SMT 3 OCH3
M SBV 3 60 0.0044 -0.6549
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 56 57
M SBL 4 1 62
M SMT 4 CH2OH
M SBV 4 62 -0.6671 0.3239
S SKP 5
ID FL5FAHGL0005
FORMULA C34H42O23
EXACTMASS 818.2116876499999
AVERAGEMASS 818.6834799999999
SMILES O(C1CO)C(Oc(c6)cc(O)c(c63)C(C(=C(c(c4)cc(OC)c(c4OC(C(O)5)OC(C(C(O)5)O)CO)O)O3)OC(O2)C(C(C(O)C2CO)O)O)=O)C(O)C(O)C1O
M END