Mol:FL5FCAGA0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.0763 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0763 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0763 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0763 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5200 0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5200 0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9637 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9637 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9637 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9637 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5200 1.5240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5200 1.5240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4074 0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4074 0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8511 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8511 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8511 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8511 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4074 1.5240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4074 1.5240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4074 -0.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4074 -0.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2950 1.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2950 1.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2720 1.1966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2720 1.1966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8389 1.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8389 1.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8389 2.1786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8389 2.1786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2720 2.5059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2720 2.5059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2950 2.1786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2950 2.1786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4569 0.1716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4569 0.1716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5200 -0.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5200 -0.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4729 2.5446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4729 2.5446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0664 -0.9462 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.0664 -0.9462 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.6491 -0.6097 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.6491 -0.6097 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.4642 -1.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4642 -1.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6491 -1.9077 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.6491 -1.9077 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.0664 -2.2443 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.0664 -2.2443 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.2512 -1.5972 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.2512 -1.5972 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.2338 -0.3212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2338 -0.3212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2102 -1.8636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2102 -1.8636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7612 -2.0225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7612 -2.0225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9680 0.3560 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.9680 0.3560 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.4104 -0.2280 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.4104 -0.2280 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0474 0.0198 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.0474 0.0198 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.6621 0.0264 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6621 0.0264 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.2154 0.4732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.2154 0.4732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.6581 0.1790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6581 0.1790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7207 -0.2616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7207 -0.2616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4124 -0.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4124 -0.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6533 -0.1057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6533 -0.1057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4335 1.8216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4335 1.8216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9334 2.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9334 2.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5515 0.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5515 0.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3881 1.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3881 1.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0446 -1.9046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0446 -1.9046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7591 -2.3169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7591 -2.3169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 18 1 0 0 0 0 | + | 22 18 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 1 39 1 0 0 0 0 | + | 1 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 24 43 1 0 0 0 0 | + | 24 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 47 -0.975 -2.1321 | + | M SVB 3 47 -0.975 -2.1321 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 45 2.6367 0.5814 | + | M SVB 2 45 2.6367 0.5814 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 43 -3.4335 1.8216 | + | M SVB 1 43 -3.4335 1.8216 |
| − | S SKP 8 | + | S SKP 8 |
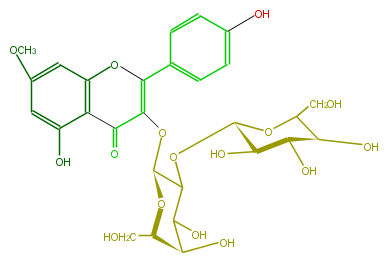
| − | ID FL5FCAGA0003 | + | ID FL5FCAGA0003 |
| − | KNApSAcK_ID C00005276 | + | KNApSAcK_ID C00005276 |
| − | NAME Rhamnocitrin 3-glucosyl-(1->2)-galactoside | + | NAME Rhamnocitrin 3-glucosyl-(1->2)-galactoside |
| − | CAS_RN 117047-50-4 | + | CAS_RN 117047-50-4 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
| − | SMILES [C@H](C1O[C@@H](O5)[C@H](O)[C@@H]([C@@H](C5CO)O)O)(OC(C(=O)2)=C(c(c4)ccc(c4)O)Oc(c3)c(c(O)cc3OC)2)O[C@H](CO)[C@H](O)C1O | + | SMILES [C@H](C1O[C@@H](O5)[C@H](O)[C@@H]([C@@H](C5CO)O)O)(OC(C(=O)2)=C(c(c4)ccc(c4)O)Oc(c3)c(c(O)cc3OC)2)O[C@H](CO)[C@H](O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-3.0763 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0763 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5200 0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9637 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9637 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5200 1.5240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4074 0.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8511 0.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8511 1.2028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4074 1.5240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4074 -0.2615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2950 1.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2720 1.1966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8389 1.5239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8389 2.1786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2720 2.5059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2950 2.1786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4569 0.1716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5200 -0.4028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4729 2.5446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0664 -0.9462 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.6491 -0.6097 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.4642 -1.2568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6491 -1.9077 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0664 -2.2443 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.2512 -1.5972 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.2338 -0.3212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2102 -1.8636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7612 -2.0225 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9680 0.3560 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.4104 -0.2280 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0474 0.0198 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.6621 0.0264 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2154 0.4732 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.6581 0.1790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7207 -0.2616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4124 -0.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6533 -0.1057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4335 1.8216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9334 2.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5515 0.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3881 1.7359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0446 -1.9046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7591 -2.3169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
8 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 18 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
31 36 1 0 0 0 0
32 37 1 0 0 0 0
27 30 1 0 0 0 0
33 38 1 0 0 0 0
1 39 1 0 0 0 0
39 40 1 0 0 0 0
34 41 1 0 0 0 0
41 42 1 0 0 0 0
24 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 47
M SMT 3 CH2OH
M SVB 3 47 -0.975 -2.1321
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 CH2OH
M SVB 2 45 2.6367 0.5814
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 OCH3
M SVB 1 43 -3.4335 1.8216
S SKP 8
ID FL5FCAGA0003
KNApSAcK_ID C00005276
NAME Rhamnocitrin 3-glucosyl-(1->2)-galactoside
CAS_RN 117047-50-4
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES [C@H](C1O[C@@H](O5)[C@H](O)[C@@H]([C@@H](C5CO)O)O)(OC(C(=O)2)=C(c(c4)ccc(c4)O)Oc(c3)c(c(O)cc3OC)2)O[C@H](CO)[C@H](O)C1O
M END