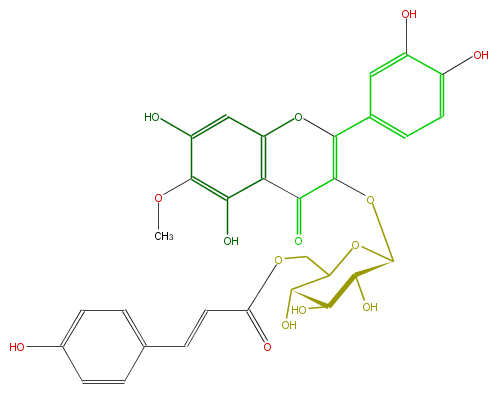
Mol:FL5FECGS0055
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.1385 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1385 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4240 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4240 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4240 2.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4240 2.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1385 2.8627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1385 2.8627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8530 2.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8530 2.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8530 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8530 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7096 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7096 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9951 1.6252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9951 1.6252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2806 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2806 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2806 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2806 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9951 -0.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9951 -0.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7096 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7096 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4338 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4338 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1483 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1483 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1483 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1483 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4338 -0.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4338 -0.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9951 -0.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9951 -0.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8628 1.6252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8628 1.6252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4240 -0.0248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4240 -0.0248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4338 -0.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4338 -0.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5675 2.8627 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5675 2.8627 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1385 3.6877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1385 3.6877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8094 0.0060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8094 0.0060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8094 -0.7571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8094 -0.7571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8343 -1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8343 -1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5864 -2.1889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5864 -2.1889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1128 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1128 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8896 -1.2746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8896 -1.2746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1375 -0.9348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1375 -0.9348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6110 -1.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6110 -1.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1418 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1418 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5904 -1.2410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5904 -1.2410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7237 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7237 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0708 -2.2402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0708 -2.2402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3468 -2.1731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3468 -2.1731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0339 -2.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0339 -2.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3792 -2.9752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3792 -2.9752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8577 -2.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8577 -2.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2697 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2697 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0936 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0936 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5061 -2.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5061 -2.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3311 -2.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3311 -2.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7436 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7436 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3311 -3.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3311 -3.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5061 -3.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5061 -3.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5675 -2.9732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5675 -2.9732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 15 23 1 0 0 0 0 | + | 15 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 28 27 1 1 0 0 0 | + | 28 27 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 26 34 1 0 0 0 0 | + | 26 34 1 0 0 0 0 |
| − | 27 35 1 0 0 0 0 | + | 27 35 1 0 0 0 0 |
| − | 28 19 1 0 0 0 0 | + | 28 19 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 43 46 1 0 0 0 0 | + | 43 46 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FECGS0055 | + | ID FL5FECGS0055 |
| − | KNApSAcK_ID C00013944 | + | KNApSAcK_ID C00013944 |
| − | NAME Patuletin 3-(6''-p-coumaroylglucoside);Quercetagetin 6-methyl ether 3-(6''-p-coumaroylglucoside);2-(3,4-Dihydroxyphenyl)-5,7-dihydroxy-3-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-6-methoxy-4H-1-benzopyran-4-one | + | NAME Patuletin 3-(6''-p-coumaroylglucoside);Quercetagetin 6-methyl ether 3-(6''-p-coumaroylglucoside);2-(3,4-Dihydroxyphenyl)-5,7-dihydroxy-3-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-6-methoxy-4H-1-benzopyran-4-one |
| − | CAS_RN 235741-39-6 | + | CAS_RN 235741-39-6 |
| − | FORMULA C31H28O15 | + | FORMULA C31H28O15 |
| − | EXACTMASS 640.1428202259999 | + | EXACTMASS 640.1428202259999 |
| − | AVERAGEMASS 640.54502 | + | AVERAGEMASS 640.54502 |
| − | SMILES C(c(c5)ccc(O)c5)=CC(OCC(O1)C(O)C(O)C(C(OC(=C3c(c4)ccc(c4O)O)C(=O)c(c2O)c(O3)cc(c(OC)2)O)1)O)=O | + | SMILES C(c(c5)ccc(O)c5)=CC(OCC(O1)C(O)C(O)C(C(OC(=C3c(c4)ccc(c4O)O)C(=O)c(c2O)c(O3)cc(c(OC)2)O)1)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
3.1385 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4240 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4240 2.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1385 2.8627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8530 2.4502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8530 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7096 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9951 1.6252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2806 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2806 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9951 -0.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7096 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4338 1.6252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1483 1.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1483 0.3877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4338 -0.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9951 -0.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8628 1.6252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4240 -0.0248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4338 -0.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5675 2.8627 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1385 3.6877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8094 0.0060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8094 -0.7571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8343 -1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5864 -2.1889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1128 -1.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8896 -1.2746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1375 -0.9348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6110 -1.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1418 -1.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5904 -1.2410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7237 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0708 -2.2402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3468 -2.1731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0339 -2.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3792 -2.9752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8577 -2.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2697 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0936 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5061 -2.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3311 -2.2587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7436 -2.9732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3311 -3.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5061 -3.6877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5675 -2.9732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
15 23 1 0 0 0 0
23 24 1 0 0 0 0
25 26 1 1 0 0 0
26 27 1 1 0 0 0
28 27 1 1 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
30 25 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
25 33 1 0 0 0 0
26 34 1 0 0 0 0
27 35 1 0 0 0 0
28 19 1 0 0 0 0
36 37 2 0 0 0 0
36 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
45 40 1 0 0 0 0
43 46 1 0 0 0 0
32 36 1 0 0 0 0
S SKP 8
ID FL5FECGS0055
KNApSAcK_ID C00013944
NAME Patuletin 3-(6''-p-coumaroylglucoside);Quercetagetin 6-methyl ether 3-(6''-p-coumaroylglucoside);2-(3,4-Dihydroxyphenyl)-5,7-dihydroxy-3-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-6-methoxy-4H-1-benzopyran-4-one
CAS_RN 235741-39-6
FORMULA C31H28O15
EXACTMASS 640.1428202259999
AVERAGEMASS 640.54502
SMILES C(c(c5)ccc(O)c5)=CC(OCC(O1)C(O)C(O)C(C(OC(=C3c(c4)ccc(c4O)O)C(=O)c(c2O)c(O3)cc(c(OC)2)O)1)O)=O
M END