Mol:FL5FFCGSS001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.8271 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8271 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8271 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8271 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2708 -1.5468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2708 -1.5468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7145 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7145 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7145 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7145 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2708 -0.2620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2708 -0.2620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1582 -1.5468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1582 -1.5468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3981 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3981 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3981 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3981 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1582 -0.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1582 -0.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1582 -2.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1582 -2.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0986 -0.1384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0986 -0.1384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6656 -0.4658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6656 -0.4658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2326 -0.1384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2326 -0.1384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2326 0.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2326 0.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6656 0.8436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6656 0.8436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0986 0.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0986 0.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2708 -2.1889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2708 -2.1889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8394 0.8666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8394 0.8666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4916 -0.1996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4916 -0.1996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6656 1.4981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6656 1.4981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2149 -1.8072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2149 -1.8072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5933 -1.8072 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5933 -1.8072 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5933 -1.3253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5933 -1.3253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0777 -1.8072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0777 -1.8072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5933 -2.3574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5933 -2.3574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2869 2.0077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2869 2.0077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5257 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5257 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0784 1.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0784 1.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8157 0.6384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8157 0.6384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6785 1.1504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6785 1.1504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1260 1.4320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1260 1.4320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8008 2.3043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8008 2.3043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8394 1.9283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8394 1.9283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3705 0.5600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3705 0.5600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2682 0.3223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2682 0.3223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9975 2.3574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9975 2.3574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2006 2.1438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2006 2.1438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 23 26 2 0 0 0 0 | + | 23 26 2 0 0 0 0 |
| − | 25 8 1 0 0 0 0 | + | 25 8 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 36 6 1 0 0 0 0 | + | 36 6 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 40 -7.9232 8.5715 | + | M SBV 1 40 -7.9232 8.5715 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FFCGSS001 | + | ID FL5FFCGSS001 |
| − | KNApSAcK_ID C00006075 | + | KNApSAcK_ID C00006075 |
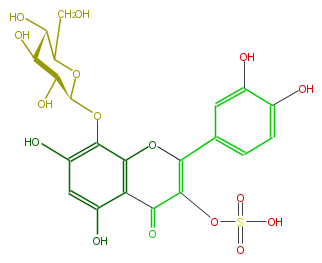
| − | NAME Gossypetin 8-glucoside-3-sulfate | + | NAME Gossypetin 8-glucoside-3-sulfate |
| − | CAS_RN 63109-35-3 | + | CAS_RN 63109-35-3 |
| − | FORMULA C21H20O16S | + | FORMULA C21H20O16S |
| − | EXACTMASS 560.047205282 | + | EXACTMASS 560.047205282 |
| − | AVERAGEMASS 560.4399 | + | AVERAGEMASS 560.4399 |
| − | SMILES c(O)(c4)c(c(c(c4O)3)OC(=C(OS(O)(=O)=O)C3=O)c(c2)cc(c(O)c2)O)OC(C(O)1)OC(CO)C(O)C1O | + | SMILES c(O)(c4)c(c(c(c4O)3)OC(=C(OS(O)(=O)=O)C3=O)c(c2)cc(c(O)c2)O)OC(C(O)1)OC(CO)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-1.8271 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8271 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2708 -1.5468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7145 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7145 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2708 -0.2620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1582 -1.5468 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3981 -1.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3981 -0.5832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1582 -0.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1582 -2.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0986 -0.1384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6656 -0.4658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2326 -0.1384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2326 0.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6656 0.8436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0986 0.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2708 -2.1889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8394 0.8666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4916 -0.1996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6656 1.4981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2149 -1.8072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5933 -1.8072 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
1.5933 -1.3253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0777 -1.8072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5933 -2.3574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2869 2.0077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5257 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0784 1.0823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8157 0.6384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6785 1.1504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1260 1.4320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8008 2.3043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8394 1.9283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3705 0.5600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2682 0.3223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9975 2.3574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2006 2.1438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 2 0 0 0 0
23 25 1 0 0 0 0
23 26 2 0 0 0 0
25 8 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
27 33 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
36 6 1 0 0 0 0
32 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 40
M SMT 1 CH2OH
M SBV 1 40 -7.9232 8.5715
S SKP 8
ID FL5FFCGSS001
KNApSAcK_ID C00006075
NAME Gossypetin 8-glucoside-3-sulfate
CAS_RN 63109-35-3
FORMULA C21H20O16S
EXACTMASS 560.047205282
AVERAGEMASS 560.4399
SMILES c(O)(c4)c(c(c(c4O)3)OC(=C(OS(O)(=O)=O)C3=O)c(c2)cc(c(O)c2)O)OC(C(O)1)OC(CO)C(O)C1O
M END