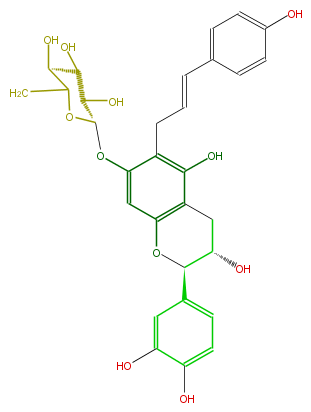
Mol:FL63ACGN0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6879 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6879 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6879 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6879 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1316 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1316 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4247 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4247 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4247 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4247 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1316 1.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1316 1.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9810 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9810 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5373 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5373 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5373 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5373 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9810 1.5466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9810 1.5466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0934 1.5465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0934 1.5465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6604 1.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6604 1.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2274 1.5465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2274 1.5465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2274 2.2012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2274 2.2012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6604 2.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6604 2.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0934 2.2012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0934 2.2012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2440 1.5465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2440 1.5465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7942 2.5285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7942 2.5285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1316 -0.3802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1316 -0.3802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0934 0.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0934 0.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2442 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2442 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2442 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2442 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6604 3.1830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6604 3.1830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8003 -0.7015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8003 -0.7015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8003 -1.3423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8003 -1.3423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3318 -1.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3318 -1.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3318 -2.2629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3318 -2.2629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8003 -2.5698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8003 -2.5698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2687 -2.2629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2687 -2.2629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2687 -1.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2687 -1.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8003 -3.1830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8003 -3.1830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2803 1.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2803 1.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9092 1.1038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9092 1.1038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3747 1.3116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3747 1.3116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8589 1.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8589 1.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2337 1.6920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2337 1.6920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7013 1.4452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7013 1.4452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7942 1.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7942 1.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4878 1.0756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4878 1.0756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0684 0.7975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0684 0.7975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0998 2.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0998 2.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3029 2.0159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3029 2.0159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 8 20 1 1 0 0 0 | + | 8 20 1 1 0 0 0 |
| − | 2 21 1 0 0 0 0 | + | 2 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 15 23 1 0 0 0 0 | + | 15 23 1 0 0 0 0 |
| − | 22 24 2 0 0 0 0 | + | 22 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 2 0 0 0 0 | + | 29 30 2 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 17 1 0 0 0 0 | + | 35 17 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 -4.8153 6.0315 | + | M SBV 1 45 -4.8153 6.0315 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL63ACGN0001 | + | ID FL63ACGN0001 |
| − | KNApSAcK_ID C00008967 | + | KNApSAcK_ID C00008967 |
| − | NAME Sachaliside 2 | + | NAME Sachaliside 2 |
| − | CAS_RN 132185-21-8 | + | CAS_RN 132185-21-8 |
| − | FORMULA C30H32O12 | + | FORMULA C30H32O12 |
| − | EXACTMASS 584.189376488 | + | EXACTMASS 584.189376488 |
| − | AVERAGEMASS 584.5678800000001 | + | AVERAGEMASS 584.5678800000001 |
| − | SMILES c(c1C=CCc(c5O)c(cc(c53)OC(c(c4)ccc(c4O)O)C(O)C3)OC(C(O)2)OC(CO)C(C(O)2)O)cc(O)cc1 | + | SMILES c(c1C=CCc(c5O)c(cc(c53)OC(c(c4)ccc(c4O)O)C(O)C3)OC(C(O)2)OC(CO)C(C(O)2)O)cc(O)cc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.6879 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6879 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1316 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4247 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4247 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1316 1.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9810 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5373 0.5831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5373 1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9810 1.5466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0934 1.5465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6604 1.2192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2274 1.5465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2274 2.2012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6604 2.5285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0934 2.2012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2440 1.5465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7942 2.5285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1316 -0.3802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0934 0.2620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2442 0.2619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2442 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6604 3.1830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8003 -0.7015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8003 -1.3423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3318 -1.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3318 -2.2629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8003 -2.5698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2687 -2.2629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2687 -1.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8003 -3.1830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2803 1.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9092 1.1038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3747 1.3116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8589 1.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2337 1.6920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7013 1.4452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7942 1.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4878 1.0756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0684 0.7975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0998 2.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3029 2.0159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 6 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
8 20 1 1 0 0 0
2 21 1 0 0 0 0
21 22 1 0 0 0 0
15 23 1 0 0 0 0
22 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 29 1 0 0 0 0
29 30 2 0 0 0 0
30 25 1 0 0 0 0
28 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 17 1 0 0 0 0
37 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 -4.8153 6.0315
S SKP 8
ID FL63ACGN0001
KNApSAcK_ID C00008967
NAME Sachaliside 2
CAS_RN 132185-21-8
FORMULA C30H32O12
EXACTMASS 584.189376488
AVERAGEMASS 584.5678800000001
SMILES c(c1C=CCc(c5O)c(cc(c53)OC(c(c4)ccc(c4O)O)C(O)C3)OC(C(O)2)OC(CO)C(C(O)2)O)cc(O)cc1
M END