Mol:FL7AAAGL0072
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 68 74 0 0 0 0 0 0 0 0999 V2000 | + | 68 74 0 0 0 0 0 0 0 0999 V2000 |
| − | 2.1001 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1001 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1001 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1001 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8146 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8146 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5290 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5290 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5290 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5290 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8146 1.6332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8146 1.6332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2435 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2435 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9580 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9580 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9580 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9580 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2435 1.6332 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 4.2435 1.6332 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7345 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7345 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4490 1.2565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4490 1.2565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.1635 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1635 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.1635 2.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1635 2.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4490 2.9065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4490 2.9065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7345 2.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7345 2.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.8037 2.8636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.8037 2.8636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5201 1.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5201 1.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8146 -0.7795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8146 -0.7795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6082 0.0203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6082 0.0203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1330 1.6617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1330 1.6617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7203 0.9470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7203 0.9470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0780 1.1562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0780 1.1562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8715 0.9293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8715 0.9293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4589 1.6441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4589 1.6441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3395 1.4350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3395 1.4350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4964 2.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4964 2.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6162 1.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6162 1.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1690 1.2061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1690 1.2061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0935 0.5162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0935 0.5162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5195 -1.7944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5195 -1.7944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2147 -2.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2147 -2.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8273 -1.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8273 -1.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.6362 -1.5227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.6362 -1.5227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9411 -1.0778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9411 -1.0778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3284 -1.6307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3284 -1.6307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.2855 -2.1882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2855 -2.1882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2864 -2.5288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2864 -2.5288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7829 -2.3340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7829 -2.3340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8519 -1.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8519 -1.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0585 2.3132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0585 2.3132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4705 3.0267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4705 3.0267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0585 3.7402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0585 3.7402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3257 3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3257 3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7382 3.7701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7382 3.7701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5632 3.7701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5632 3.7701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9757 3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9757 3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5632 2.3412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5632 2.3412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7382 2.3412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7382 2.3412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7996 3.0557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7996 3.0557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.3205 2.9652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.3205 2.9652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.9078 2.2505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.9078 2.2505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1095 2.4597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1095 2.4597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3160 2.2328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3160 2.2328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7286 2.9476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7286 2.9476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5270 2.7385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5270 2.7385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6839 3.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6839 3.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.1638 3.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.1638 3.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.8037 2.4820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.8037 2.4820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.3565 2.5096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.3565 2.5096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2810 1.8197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2810 1.8197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7544 -3.7701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7544 -3.7701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7670 -2.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7670 -2.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0599 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0599 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4867 -2.5454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4867 -2.5454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4994 -1.7216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4994 -1.7216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7922 -1.2988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7922 -1.2988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2191 -1.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2191 -1.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 44 1 0 0 0 0 | + | 49 44 1 0 0 0 0 |
| − | 47 50 1 0 0 0 0 | + | 47 50 1 0 0 0 0 |
| − | 41 27 1 0 0 0 0 | + | 41 27 1 0 0 0 0 |
| − | 44 42 1 0 0 0 0 | + | 44 42 1 0 0 0 0 |
| − | 51 52 1 1 0 0 0 | + | 51 52 1 1 0 0 0 |
| − | 52 53 1 1 0 0 0 | + | 52 53 1 1 0 0 0 |
| − | 54 53 1 1 0 0 0 | + | 54 53 1 1 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 56 51 1 0 0 0 0 | + | 56 51 1 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | 57 58 1 0 0 0 0 | + | 57 58 1 0 0 0 0 |
| − | 51 59 1 0 0 0 0 | + | 51 59 1 0 0 0 0 |
| − | 52 60 1 0 0 0 0 | + | 52 60 1 0 0 0 0 |
| − | 53 61 1 0 0 0 0 | + | 53 61 1 0 0 0 0 |
| − | 50 54 1 0 0 0 0 | + | 50 54 1 0 0 0 0 |
| − | 62 63 2 0 0 0 0 | + | 62 63 2 0 0 0 0 |
| − | 63 64 1 0 0 0 0 | + | 63 64 1 0 0 0 0 |
| − | 63 65 1 0 0 0 0 | + | 63 65 1 0 0 0 0 |
| − | 65 66 1 0 0 0 0 | + | 65 66 1 0 0 0 0 |
| − | 66 67 2 0 0 0 0 | + | 66 67 2 0 0 0 0 |
| − | 66 68 1 0 0 0 0 | + | 66 68 1 0 0 0 0 |
| − | 68 40 1 0 0 0 0 | + | 68 40 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0072 | + | ID FL7AAAGL0072 |
| − | KNApSAcK_ID C00014851 | + | KNApSAcK_ID C00014851 |
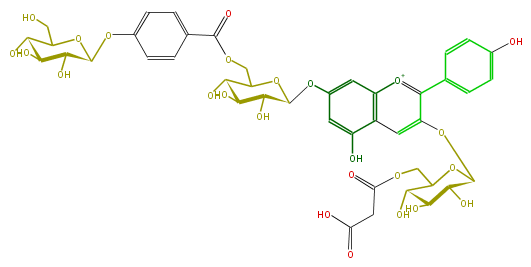
| − | NAME Pelargonidin 3-(6-(malonyl)glucoside)-7-(6-(4-(glucosyl)-p-hydroxybenzoyl)glucoside) | + | NAME Pelargonidin 3-(6-(malonyl)glucoside)-7-(6-(4-(glucosyl)-p-hydroxybenzoyl)glucoside) |
| − | CAS_RN 215237-92-6 | + | CAS_RN 215237-92-6 |
| − | FORMULA C43H47O25 | + | FORMULA C43H47O25 |
| − | EXACTMASS 963.2406420539999 | + | EXACTMASS 963.2406420539999 |
| − | AVERAGEMASS 963.8182800000001 | + | AVERAGEMASS 963.8182800000001 |
| − | SMILES c(c6)(c(c(c7)ccc(c7)O)[o+1]c(c26)cc(OC(C(O)3)OC(COC(c(c5)ccc(c5)OC(C(O)4)OC(CO)C(C(O)4)O)=O)C(O)C(O)3)cc2O)OC(O1)C(C(O)C(C1COC(=O)CC(O)=O)O)O | + | SMILES c(c6)(c(c(c7)ccc(c7)O)[o+1]c(c26)cc(OC(C(O)3)OC(COC(c(c5)ccc(c5)OC(C(O)4)OC(CO)C(C(O)4)O)=O)C(O)C(O)3)cc2O)OC(O1)C(C(O)C(C1COC(=O)CC(O)=O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
68 74 0 0 0 0 0 0 0 0999 V2000
2.1001 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1001 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8146 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5290 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5290 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8146 1.6332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2435 -0.0168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9580 0.3957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9580 1.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2435 1.6332 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
5.7345 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4490 1.2565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.1635 1.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.1635 2.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4490 2.9065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7345 2.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.8037 2.8636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5201 1.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8146 -0.7795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6082 0.0203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1330 1.6617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7203 0.9470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0780 1.1562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8715 0.9293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4589 1.6441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3395 1.4350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4964 2.0208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6162 1.1785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1690 1.2061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0935 0.5162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5195 -1.7944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2147 -2.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8273 -1.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.6362 -1.5227 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9411 -1.0778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3284 -1.6307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2855 -2.1882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2864 -2.5288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7829 -2.3340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8519 -1.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0585 2.3132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4705 3.0267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0585 3.7402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3257 3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7382 3.7701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5632 3.7701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9757 3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5632 2.3412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7382 2.3412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7996 3.0557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-7.3205 2.9652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.9078 2.2505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1095 2.4597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3160 2.2328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7286 2.9476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.5270 2.7385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.6839 3.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.1638 3.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-7.8037 2.4820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-7.3565 2.5096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.2810 1.8197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7544 -3.7701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7670 -2.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0599 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4867 -2.5454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4994 -1.7216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7922 -1.2988 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2191 -1.3206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
8 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
18 24 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
33 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
36 40 1 0 0 0 0
34 20 1 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 44 1 0 0 0 0
47 50 1 0 0 0 0
41 27 1 0 0 0 0
44 42 1 0 0 0 0
51 52 1 1 0 0 0
52 53 1 1 0 0 0
54 53 1 1 0 0 0
54 55 1 0 0 0 0
55 56 1 0 0 0 0
56 51 1 0 0 0 0
56 57 1 0 0 0 0
57 58 1 0 0 0 0
51 59 1 0 0 0 0
52 60 1 0 0 0 0
53 61 1 0 0 0 0
50 54 1 0 0 0 0
62 63 2 0 0 0 0
63 64 1 0 0 0 0
63 65 1 0 0 0 0
65 66 1 0 0 0 0
66 67 2 0 0 0 0
66 68 1 0 0 0 0
68 40 1 0 0 0 0
S SKP 8
ID FL7AAAGL0072
KNApSAcK_ID C00014851
NAME Pelargonidin 3-(6-(malonyl)glucoside)-7-(6-(4-(glucosyl)-p-hydroxybenzoyl)glucoside)
CAS_RN 215237-92-6
FORMULA C43H47O25
EXACTMASS 963.2406420539999
AVERAGEMASS 963.8182800000001
SMILES c(c6)(c(c(c7)ccc(c7)O)[o+1]c(c26)cc(OC(C(O)3)OC(COC(c(c5)ccc(c5)OC(C(O)4)OC(CO)C(C(O)4)O)=O)C(O)C(O)3)cc2O)OC(O1)C(C(O)C(C1COC(=O)CC(O)=O)O)O
M END