Mol:FL7AAGGL0030
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 64 69 0 0 0 0 0 0 0 0999 V2000 | + | 64 69 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.8350 -0.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8350 -0.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2246 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2246 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2246 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2246 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8350 1.1550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8350 1.1550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4454 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4454 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4454 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4454 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6141 -0.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6141 -0.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0037 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0037 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0037 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0037 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6141 1.1550 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.6141 1.1550 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5778 1.1382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5778 1.1382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1614 0.8013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1614 0.8013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7450 1.1382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7450 1.1382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7450 1.8121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7450 1.8121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1614 2.1490 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1614 2.1490 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5778 1.8121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5778 1.8121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3511 2.1620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3511 2.1620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9448 1.0908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9448 1.0908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8350 -0.9153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8350 -0.9153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4338 -0.3373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4338 -0.3373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1763 2.6987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1763 2.6987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3808 0.8507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3808 0.8507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3947 -1.1416 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.3947 -1.1416 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.8792 -1.7857 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.8792 -1.7857 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.1218 -1.4587 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.1218 -1.4587 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3034 -1.5633 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.3034 -1.5633 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.8189 -0.9191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8189 -0.9191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5764 -1.2460 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.5764 -1.2460 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.8507 -1.5453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8507 -1.5453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3475 -1.6292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3475 -1.6292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2846 -1.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2846 -1.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4972 -0.3406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4972 -0.3406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0785 -0.2897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0785 -0.2897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1865 0.3223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1865 0.3223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7943 0.8824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7943 0.8824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7960 0.3757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7960 0.3757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0457 0.9112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0457 0.9112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6851 1.4262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6851 1.4262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5748 0.9575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5748 0.9575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4526 -0.4530 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.4526 -0.4530 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9037 -1.0690 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9037 -1.0690 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.7264 -1.0079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7264 -1.0079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4654 -1.3747 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4654 -1.3747 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0144 -0.7588 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.0144 -0.7588 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1916 -0.8197 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.1916 -0.8197 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.0033 -0.5734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0033 -0.5734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6364 -0.3750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6364 -0.3750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0038 -0.6134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0038 -0.6134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4654 -1.3747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4654 -1.3747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9536 -1.7422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9536 -1.7422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4271 -2.2157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4271 -2.2157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8778 -1.7650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8778 -1.7650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4271 -2.6987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4271 -2.6987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3812 -2.2684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3812 -2.2684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8950 -1.7546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8950 -1.7546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4666 -2.0847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4666 -2.0847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0383 -1.7546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0383 -1.7546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0383 -1.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0383 -1.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4666 -0.7645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4666 -0.7645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8950 -1.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8950 -1.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4773 -0.8410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4773 -0.8410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4604 -1.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4604 -1.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1317 -1.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1317 -1.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4604 -2.5816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4604 -2.5816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 5 18 1 0 0 0 0 | + | 5 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 13 22 1 0 0 0 0 | + | 13 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 19 26 1 0 0 0 0 | + | 19 26 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 20 41 1 0 0 0 0 | + | 20 41 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 44 48 1 0 0 0 0 | + | 44 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 51 53 2 0 0 0 0 | + | 51 53 2 0 0 0 0 |
| − | 52 54 2 0 0 0 0 | + | 52 54 2 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 55 56 2 0 0 0 0 | + | 55 56 2 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | 57 58 2 0 0 0 0 | + | 57 58 2 0 0 0 0 |
| − | 58 59 1 0 0 0 0 | + | 58 59 1 0 0 0 0 |
| − | 59 60 2 0 0 0 0 | + | 59 60 2 0 0 0 0 |
| − | 60 55 1 0 0 0 0 | + | 60 55 1 0 0 0 0 |
| − | 58 61 1 0 0 0 0 | + | 58 61 1 0 0 0 0 |
| − | 29 62 1 0 0 0 0 | + | 29 62 1 0 0 0 0 |
| − | 62 63 1 0 0 0 0 | + | 62 63 1 0 0 0 0 |
| − | 62 64 2 0 0 0 0 | + | 62 64 2 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 37 38 39 | + | M SAL 1 3 37 38 39 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SVB 1 40 -6.0173 -0.3217 | + | M SVB 1 40 -6.0173 -0.3217 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAGGL0030 | + | ID FL7AAGGL0030 |
| − | KNApSAcK_ID C00011082 | + | KNApSAcK_ID C00011082 |
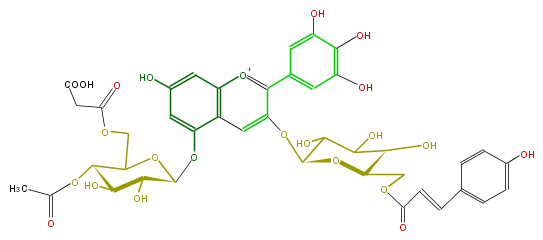
| − | NAME Acetylmalonylawobanin;Delphinidin 3-O-[6-O-(p-coumaroyl)-beta-D-glucopyranoside]-5-O-[4-O-acetyl-6-O-malonyl-beta-D-glucopyranoside] | + | NAME Acetylmalonylawobanin;Delphinidin 3-O-[6-O-(p-coumaroyl)-beta-D-glucopyranoside]-5-O-[4-O-acetyl-6-O-malonyl-beta-D-glucopyranoside] |
| − | CAS_RN 250344-52-6 | + | CAS_RN 250344-52-6 |
| − | FORMULA C41H41O23 | + | FORMULA C41H41O23 |
| − | EXACTMASS 901.203862618 | + | EXACTMASS 901.203862618 |
| − | AVERAGEMASS 901.75044 | + | AVERAGEMASS 901.75044 |
| − | SMILES Oc(c5)cc(O[C@@H]([C@H]6O)OC(COC(=O)CC(O)=O)[C@H](OC(C)=O)[C@H](O)6)c(c25)cc(O[C@H](O3)C(O)C([C@H]([C@H]3COC(=O)C=Cc(c4)ccc(O)c4)O)O)c([o+1]2)c(c1)cc(O)c(O)c(O)1 | + | SMILES Oc(c5)cc(O[C@@H]([C@H]6O)OC(COC(=O)CC(O)=O)[C@H](OC(C)=O)[C@H](O)6)c(c25)cc(O[C@H](O3)C(O)C([C@H]([C@H]3COC(=O)C=Cc(c4)ccc(O)c4)O)O)c([o+1]2)c(c1)cc(O)c(O)c(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
64 69 0 0 0 0 0 0 0 0999 V2000
-1.8350 -0.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2246 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2246 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8350 1.1550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4454 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4454 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6141 -0.2548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0037 0.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0037 0.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6141 1.1550 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.5778 1.1382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1614 0.8013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7450 1.1382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7450 1.8121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1614 2.1490 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5778 1.8121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3511 2.1620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9448 1.0908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8350 -0.9153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4338 -0.3373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1763 2.6987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3808 0.8507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3947 -1.1416 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.8792 -1.7857 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.1218 -1.4587 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3034 -1.5633 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8189 -0.9191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5764 -1.2460 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.8507 -1.5453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3475 -1.6292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2846 -1.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4972 -0.3406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0785 -0.2897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1865 0.3223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7943 0.8824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7960 0.3757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0457 0.9112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6851 1.4262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5748 0.9575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4526 -0.4530 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9037 -1.0690 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.7264 -1.0079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4654 -1.3747 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0144 -0.7588 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1916 -0.8197 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.0033 -0.5734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6364 -0.3750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0038 -0.6134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4654 -1.3747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9536 -1.7422 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4271 -2.2157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8778 -1.7650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4271 -2.6987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3812 -2.2684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8950 -1.7546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4666 -2.0847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0383 -1.7546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0383 -1.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4666 -0.7645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8950 -1.0945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4773 -0.8410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4604 -1.8973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1317 -1.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4604 -2.5816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
2 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 3 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
5 18 1 0 0 0 0
1 19 1 0 0 0 0
8 20 1 0 0 0 0
15 21 1 0 0 0 0
13 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
19 26 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
28 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
34 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
37 39 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 40 1 0 0 0 0
20 41 1 0 0 0 0
40 46 1 0 0 0 0
45 47 1 0 0 0 0
44 48 1 0 0 0 0
43 49 1 0 0 0 0
49 50 1 0 0 0 0
50 51 1 0 0 0 0
51 52 1 0 0 0 0
51 53 2 0 0 0 0
52 54 2 0 0 0 0
54 55 1 0 0 0 0
55 56 2 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
58 59 1 0 0 0 0
59 60 2 0 0 0 0
60 55 1 0 0 0 0
58 61 1 0 0 0 0
29 62 1 0 0 0 0
62 63 1 0 0 0 0
62 64 2 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 37 38 39
M SBL 1 1 40
M SMT 1 COOH
M SVB 1 40 -6.0173 -0.3217
S SKP 8
ID FL7AAGGL0030
KNApSAcK_ID C00011082
NAME Acetylmalonylawobanin;Delphinidin 3-O-[6-O-(p-coumaroyl)-beta-D-glucopyranoside]-5-O-[4-O-acetyl-6-O-malonyl-beta-D-glucopyranoside]
CAS_RN 250344-52-6
FORMULA C41H41O23
EXACTMASS 901.203862618
AVERAGEMASS 901.75044
SMILES Oc(c5)cc(O[C@@H]([C@H]6O)OC(COC(=O)CC(O)=O)[C@H](OC(C)=O)[C@H](O)6)c(c25)cc(O[C@H](O3)C(O)C([C@H]([C@H]3COC(=O)C=Cc(c4)ccc(O)c4)O)O)c([o+1]2)c(c1)cc(O)c(O)c(O)1
M END