Mol:FLIF1LGF0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 46 0 0 0 0 0 0 0 0999 V2000 | + | 41 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.1595 2.1363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1595 2.1363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9426 2.0125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9426 2.0125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4325 0.9637 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.4325 0.9637 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.9041 0.7410 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.9041 0.7410 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5692 1.1583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5692 1.1583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1163 1.4131 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1163 1.4131 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.6328 1.5313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6328 1.5313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9824 1.1183 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.9824 1.1183 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.0477 0.3485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0477 0.3485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3584 0.4484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3584 0.4484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0820 0.8858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0820 0.8858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5112 0.0177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5112 0.0177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0451 0.3389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0451 0.3389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6012 0.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6012 0.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6012 -0.6481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6012 -0.6481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1779 -0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1779 -0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7546 -0.6481 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.7546 -0.6481 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.7546 0.0178 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.7546 0.0178 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.1779 0.3508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1779 0.3508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5110 -0.6243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5110 -0.6243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0451 -0.9454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0451 -0.9454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1779 -1.6466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1779 -1.6466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3310 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3310 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3310 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3310 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8823 -1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8823 -1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4337 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4337 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4337 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4337 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8823 -0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8823 -0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5605 2.3890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5605 2.3890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0885 0.9672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0885 0.9672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7273 1.0344 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.7273 1.0344 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.9886 0.4475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9886 0.4475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9081 0.0178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9081 0.0178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3314 0.3508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3314 0.3508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6026 2.5075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6026 2.5075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5573 1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5573 1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7005 2.4635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7005 2.4635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4337 -1.6175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4337 -1.6175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4337 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4337 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4435 -2.0950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4435 -2.0950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1580 -2.5075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1580 -2.5075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3 4 1 1 0 0 0 | + | 3 4 1 1 0 0 0 |
| − | 4 5 1 1 0 0 0 | + | 4 5 1 1 0 0 0 |
| − | 6 5 1 1 0 0 0 | + | 6 5 1 1 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 3 1 0 0 0 0 | + | 8 3 1 0 0 0 0 |
| − | 3 9 1 0 0 0 0 | + | 3 9 1 0 0 0 0 |
| − | 4 10 1 0 0 0 0 | + | 4 10 1 0 0 0 0 |
| − | 5 11 1 0 0 0 0 | + | 5 11 1 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 12 20 1 0 0 0 0 | + | 12 20 1 0 0 0 0 |
| − | 20 21 2 0 0 0 0 | + | 20 21 2 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 16 22 2 0 0 0 0 | + | 16 22 2 0 0 0 0 |
| − | 17 23 1 0 0 0 0 | + | 17 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 2 29 2 0 0 0 0 | + | 2 29 2 0 0 0 0 |
| − | 13 30 1 0 0 0 0 | + | 13 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 12 1 0 0 0 0 | + | 32 12 1 0 0 0 0 |
| − | 28 33 1 0 0 0 0 | + | 28 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 18 1 0 0 0 0 | + | 34 18 1 0 0 0 0 |
| − | 31 2 1 0 0 0 0 | + | 31 2 1 0 0 0 0 |
| − | 2 35 1 0 0 0 0 | + | 2 35 1 0 0 0 0 |
| − | 35 1 1 0 0 0 0 | + | 35 1 1 0 0 0 0 |
| − | 8 36 1 0 0 0 0 | + | 8 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 26 38 1 0 0 0 0 | + | 26 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 36 37 | + | M SAL 3 2 36 37 |
| − | M SBL 3 1 41 | + | M SBL 3 1 41 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 41 -3.7589 1.225 | + | M SVB 3 41 -3.7589 1.225 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 45 2.4435 -2.095 | + | M SVB 2 45 2.4435 -2.095 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 43 3.6278 -1.8238 | + | M SVB 1 43 3.6278 -1.8238 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLIF1LGF0001 | + | ID FLIF1LGF0001 |
| − | KNApSAcK_ID C00010176 | + | KNApSAcK_ID C00010176 |
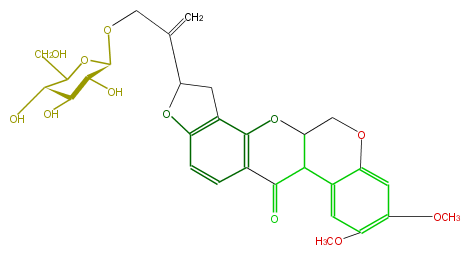
| − | NAME Amorphigenin O-glucoside | + | NAME Amorphigenin O-glucoside |
| − | CAS_RN 23708-98-7 | + | CAS_RN 23708-98-7 |
| − | FORMULA C29H32O12 | + | FORMULA C29H32O12 |
| − | EXACTMASS 572.189376488 | + | EXACTMASS 572.189376488 |
| − | AVERAGEMASS 572.55718 | + | AVERAGEMASS 572.55718 |
| − | SMILES C(O)C(O1)[C@@H]([C@H](O)[C@H](O)[C@@H]1OCC(C(O6)Cc(c65)c(c2cc5)OC(C3)C(c(c4)c(cc(OC)c4OC)O3)C2=O)=C)O | + | SMILES C(O)C(O1)[C@@H]([C@H](O)[C@H](O)[C@@H]1OCC(C(O6)Cc(c65)c(c2cc5)OC(C3)C(c(c4)c(cc(OC)c4OC)O3)C2=O)=C)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 46 0 0 0 0 0 0 0 0999 V2000
-2.1595 2.1363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9426 2.0125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4325 0.9637 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.9041 0.7410 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5692 1.1583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1163 1.4131 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.6328 1.5313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9824 1.1183 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.0477 0.3485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3584 0.4484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0820 0.8858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5112 0.0177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0451 0.3389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6012 0.0178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6012 -0.6481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1779 -0.9811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7546 -0.6481 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.7546 0.0178 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.1779 0.3508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5110 -0.6243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0451 -0.9454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1779 -1.6466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3310 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3310 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8823 -1.9358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4337 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4337 -0.9809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8823 -0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5605 2.3890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0885 0.9672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7273 1.0344 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.9886 0.4475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9081 0.0178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3314 0.3508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6026 2.5075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5573 1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7005 2.4635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4337 -1.6175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4337 -1.6175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4435 -2.0950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1580 -2.5075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3 4 1 1 0 0 0
4 5 1 1 0 0 0
6 5 1 1 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 3 1 0 0 0 0
3 9 1 0 0 0 0
4 10 1 0 0 0 0
5 11 1 0 0 0 0
6 1 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 19 1 0 0 0 0
19 14 1 0 0 0 0
12 20 1 0 0 0 0
20 21 2 0 0 0 0
21 15 1 0 0 0 0
16 22 2 0 0 0 0
17 23 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
2 29 2 0 0 0 0
13 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 12 1 0 0 0 0
28 33 1 0 0 0 0
33 34 1 0 0 0 0
34 18 1 0 0 0 0
31 2 1 0 0 0 0
2 35 1 0 0 0 0
35 1 1 0 0 0 0
8 36 1 0 0 0 0
36 37 1 0 0 0 0
26 38 1 0 0 0 0
38 39 1 0 0 0 0
25 40 1 0 0 0 0
40 41 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 36 37
M SBL 3 1 41
M SMT 3 CH2OH
M SVB 3 41 -3.7589 1.225
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 45
M SMT 2 OCH3
M SVB 2 45 2.4435 -2.095
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 43
M SMT 1 OCH3
M SVB 1 43 3.6278 -1.8238
S SKP 8
ID FLIF1LGF0001
KNApSAcK_ID C00010176
NAME Amorphigenin O-glucoside
CAS_RN 23708-98-7
FORMULA C29H32O12
EXACTMASS 572.189376488
AVERAGEMASS 572.55718
SMILES C(O)C(O1)[C@@H]([C@H](O)[C@H](O)[C@@H]1OCC(C(O6)Cc(c65)c(c2cc5)OC(C3)C(c(c4)c(cc(OC)c4OC)O3)C2=O)=C)O
M END