Mol:FL3FACCS0020
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3062 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3062 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3062 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3062 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5917 -1.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5917 -1.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1228 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1228 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1228 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1228 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5917 -0.2904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5917 -0.2904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8372 -1.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8372 -1.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5517 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5517 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5517 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5517 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8372 -0.2904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8372 -0.2904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8372 -2.5837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8372 -2.5837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0204 -0.2905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0204 -0.2905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4003 2.4973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4003 2.4973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1781 1.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1781 1.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8481 1.0595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8481 1.0595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8393 0.2409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8393 0.2409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2442 0.8358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2442 0.8358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6360 1.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6360 1.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3125 2.4973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3125 2.4973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1781 2.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1781 2.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2713 0.3266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2713 0.3266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5917 -2.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5917 -2.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3453 -0.2641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3453 -0.2641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0981 -0.6988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0981 -0.6988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8511 -0.2641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8511 -0.2641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8511 0.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8511 0.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0981 1.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0981 1.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3453 0.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3453 0.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9735 -1.7399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9735 -1.7399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4968 -2.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4968 -2.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8103 -2.1022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8103 -2.1022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1480 -2.0950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1480 -2.0950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6293 -1.6135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6293 -1.6135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2298 -1.9305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2298 -1.9305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6034 -1.9087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6034 -1.9087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1484 -2.3691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1484 -2.3691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1824 -2.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1824 -2.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6034 1.0396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6034 1.0396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6034 -0.6986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6034 -0.6986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7684 -1.3918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7684 -1.3918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8509 -1.9216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8509 -1.9216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 9 1 0 0 0 0 | + | 23 9 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 2 1 0 0 0 0 | + | 32 2 1 0 0 0 0 |
| − | 26 38 1 0 0 0 0 | + | 26 38 1 0 0 0 0 |
| − | 25 39 1 0 0 0 0 | + | 25 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 0.5386 -0.5386 | + | M SBV 1 45 0.5386 -0.5386 |
| − | S SKP 5 | + | S SKP 5 |
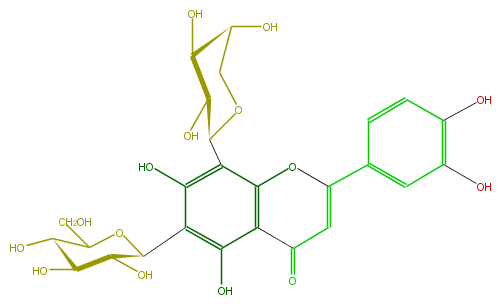
| − | ID FL3FACCS0020 | + | ID FL3FACCS0020 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O | + | SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-1.3062 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3062 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5917 -1.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1228 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1228 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5917 -0.2904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8372 -1.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5517 -1.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5517 -0.7029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8372 -0.2904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8372 -2.5837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0204 -0.2905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4003 2.4973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1781 1.9081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8481 1.0595 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8393 0.2409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2442 0.8358 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6360 1.5781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3125 2.4973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1781 2.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2713 0.3266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5917 -2.7651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3453 -0.2641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0981 -0.6988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8511 -0.2641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8511 0.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0981 1.0399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3453 0.6053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9735 -1.7399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4968 -2.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8103 -2.1022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1480 -2.0950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6293 -1.6135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2298 -1.9305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6034 -1.9087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1484 -2.3691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1824 -2.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6034 1.0396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6034 -0.6986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7684 -1.3918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8509 -1.9216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
23 9 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 2 1 0 0 0 0
26 38 1 0 0 0 0
25 39 1 0 0 0 0
40 41 1 0 0 0 0
34 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 0.5386 -0.5386
S SKP 5
ID FL3FACCS0020
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES C(C(C1O)OC(c(c4O)c(c(c(c4C(O5)C(O)C(O)C(O)C5)2)C(C=C(c(c3)ccc(O)c3O)O2)=O)O)C(C1O)O)O
M END