Mol:FL3FACDS0026
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | 4.0823 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0823 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3678 0.2762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3678 0.2762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6534 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6534 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6534 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6534 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3678 -1.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3678 -1.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0823 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0823 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9389 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9389 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2244 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2244 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2244 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2244 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9389 -1.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9389 -1.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3678 -1.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3678 -1.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8837 0.3264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8837 0.3264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5982 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5982 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3126 0.3264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3126 0.3264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3126 1.1514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3126 1.1514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5982 1.5639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5982 1.5639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8837 1.1514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8837 1.1514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.9961 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.9961 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9389 -2.1437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9389 -2.1437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5340 0.2624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5340 0.2624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.9742 -0.0555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.9742 -0.0555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9589 -1.3073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9589 -1.3073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5462 -2.0220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5462 -2.0220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7479 -1.8128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7479 -1.8128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0456 -2.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0456 -2.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3670 -1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3670 -1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1654 -1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1654 -1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3223 -0.9482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3223 -0.9482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8022 -0.6711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8022 -0.6711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4421 -1.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4421 -1.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9949 -1.7630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9949 -1.7630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9194 -2.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9194 -2.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9919 0.2932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9919 0.2932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5792 -0.4215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5792 -0.4215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7809 -0.2123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7809 -0.2123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0126 -0.4392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0126 -0.4392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4000 0.2756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4000 0.2756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1984 0.0665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1984 0.0665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3553 0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3553 0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4751 -0.1900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4751 -0.1900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0279 -0.1625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0279 -0.1625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9524 -0.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9524 -0.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9826 1.0258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9826 1.0258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3945 1.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3945 1.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9826 2.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9826 2.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2184 1.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2184 1.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6304 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6304 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4542 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4542 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8667 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8667 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6917 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6917 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1042 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1042 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6917 0.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6917 0.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8667 0.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8667 0.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.9281 1.0258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.9281 1.0258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2699 2.3184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2699 2.3184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.9961 2.3184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.9961 2.3184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 2 0 0 0 0 | + | 5 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 10 19 1 0 0 0 0 | + | 10 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 14 21 1 0 0 0 0 | + | 14 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 9 1 0 0 0 0 | + | 25 9 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 20 36 1 0 0 0 0 | + | 20 36 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 48 1 0 0 0 0 | + | 53 48 1 0 0 0 0 |
| − | 51 54 1 0 0 0 0 | + | 51 54 1 0 0 0 0 |
| − | 50 55 1 0 0 0 0 | + | 50 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 43 39 1 0 0 0 0 | + | 43 39 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACDS0026 | + | ID FL3FACDS0026 |
| − | KNApSAcK_ID C00014097 | + | KNApSAcK_ID C00014097 |
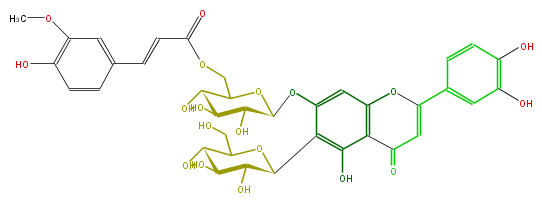
| − | NAME Isoorientin 7-O-(6'''-O-(E)-feruloyl)glucoside | + | NAME Isoorientin 7-O-(6'''-O-(E)-feruloyl)glucoside |
| − | CAS_RN 496788-50-2 | + | CAS_RN 496788-50-2 |
| − | FORMULA C37H38O19 | + | FORMULA C37H38O19 |
| − | EXACTMASS 786.200729034 | + | EXACTMASS 786.200729034 |
| − | AVERAGEMASS 786.68622 | + | AVERAGEMASS 786.68622 |
| − | SMILES COc(c(O)6)cc(cc6)C=CC(OCC(C(O)1)OC(Oc(c3)c(c(c(C(=O)4)c(OC(c(c5)ccc(O)c(O)5)=C4)3)O)C(O2)C(O)C(O)C(C2CO)O)C(O)C(O)1)=O | + | SMILES COc(c(O)6)cc(cc6)C=CC(OCC(C(O)1)OC(Oc(c3)c(c(c(C(=O)4)c(OC(c(c5)ccc(O)c(O)5)=C4)3)O)C(O2)C(O)C(O)C(C2CO)O)C(O)C(O)1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
4.0823 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3678 0.2762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6534 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6534 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3678 -1.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0823 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9389 0.2762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2244 -0.1363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2244 -0.9613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9389 -1.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3678 -1.9458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8837 0.3264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5982 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3126 0.3264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3126 1.1514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5982 1.5639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8837 1.1514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.9961 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9389 -2.1437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5340 0.2624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.9742 -0.0555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9589 -1.3073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5462 -2.0220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7479 -1.8128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0456 -2.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3670 -1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1654 -1.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3223 -0.9482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8022 -0.6711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4421 -1.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9949 -1.7630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9194 -2.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9919 0.2932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5792 -0.4215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7809 -0.2123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0126 -0.4392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4000 0.2756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1984 0.0665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3553 0.6523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4751 -0.1900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0279 -0.1625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9524 -0.8523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9826 1.0258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3945 1.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9826 2.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2184 1.7393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6304 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4542 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8667 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6917 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1042 1.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6917 0.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8667 0.3113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.9281 1.0258 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.2699 2.3184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.9961 2.3184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
3 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 2 0 0 0 0
1 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
10 19 1 0 0 0 0
8 20 1 0 0 0 0
14 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 9 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
38 39 1 0 0 0 0
33 40 1 0 0 0 0
34 41 1 0 0 0 0
35 42 1 0 0 0 0
20 36 1 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 48 1 0 0 0 0
51 54 1 0 0 0 0
50 55 1 0 0 0 0
55 56 1 0 0 0 0
43 39 1 0 0 0 0
S SKP 8
ID FL3FACDS0026
KNApSAcK_ID C00014097
NAME Isoorientin 7-O-(6'''-O-(E)-feruloyl)glucoside
CAS_RN 496788-50-2
FORMULA C37H38O19
EXACTMASS 786.200729034
AVERAGEMASS 786.68622
SMILES COc(c(O)6)cc(cc6)C=CC(OCC(C(O)1)OC(Oc(c3)c(c(c(C(=O)4)c(OC(c(c5)ccc(O)c(O)5)=C4)3)O)C(O2)C(O)C(O)C(C2CO)O)C(O)C(O)1)=O
M END