Mol:FL3FACGS0037
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.4834 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4834 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4834 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4834 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7824 -1.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7824 -1.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0815 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0815 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0815 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0815 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7824 -0.3476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7824 -0.3476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3804 -1.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3804 -1.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6794 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6794 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6794 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6794 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3804 -0.3476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3804 -0.3476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3804 -2.7374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3804 -2.7374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0213 -0.3478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0213 -0.3478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7358 -0.7604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7358 -0.7604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4502 -0.3478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4502 -0.3478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4502 0.4771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4502 0.4771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7358 0.8896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7358 0.8896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0213 0.4771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0213 0.4771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2907 -0.2862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2907 -0.2862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7824 -2.7743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7824 -2.7743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2088 0.8156 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2088 0.8156 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7358 1.7145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7358 1.7145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7626 0.9074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7626 0.9074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0792 0.5128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0792 0.5128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2960 1.2716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2960 1.2716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0792 2.0351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0792 2.0351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7626 2.4297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7626 2.4297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5458 1.6708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5458 1.6708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3428 2.2725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3428 2.2725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1397 2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1397 2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7935 0.9815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7935 0.9815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3860 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3860 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9644 0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9644 0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7753 0.9338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7753 0.9338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5911 0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5911 0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0129 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0129 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2019 1.2008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2019 1.2008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6028 1.2226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6028 1.2226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6197 1.6953 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6197 1.6953 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6215 1.0812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6215 1.0812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1483 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1483 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1483 2.7743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1483 2.7743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9255 1.4281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9255 1.4281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1483 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1483 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9255 0.2824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9255 0.2824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 18 23 1 0 0 0 0 | + | 18 23 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 32 20 1 0 0 0 0 | + | 32 20 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 27 40 1 0 0 0 0 | + | 27 40 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 34 43 1 0 0 0 0 | + | 34 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 40 41 42 | + | M SAL 1 3 40 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^COOH | + | M SMT 1 ^COOH |
| − | M SBV 1 46 0.6026 -0.2060 | + | M SBV 1 46 0.6026 -0.2060 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 48 -0.5572 -0.0290 | + | M SBV 2 48 -0.5572 -0.0290 |
| − | S SKP 5 | + | S SKP 5 |
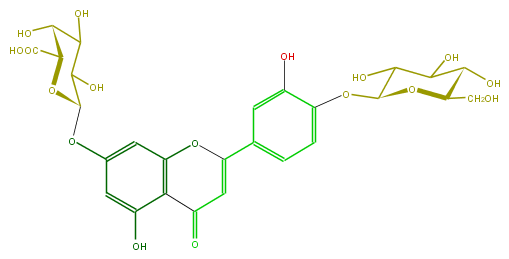
| − | ID FL3FACGS0037 | + | ID FL3FACGS0037 |
| − | FORMULA C27H28O17 | + | FORMULA C27H28O17 |
| − | EXACTMASS 624.1326494699999 | + | EXACTMASS 624.1326494699999 |
| − | AVERAGEMASS 624.50102 | + | AVERAGEMASS 624.50102 |
| − | SMILES O(c13)C(c(c4)cc(O)c(OC(O5)C(C(C(O)C(CO)5)O)O)c4)=CC(=O)c(c(cc(c3)OC(O2)C(C(C(O)C2C(O)=O)O)O)O)1 | + | SMILES O(c13)C(c(c4)cc(O)c(OC(O5)C(C(C(O)C(CO)5)O)O)c4)=CC(=O)c(c(cc(c3)OC(O2)C(C(C(O)C2C(O)=O)O)O)O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-3.4834 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4834 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7824 -1.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0815 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0815 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7824 -0.3476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3804 -1.9665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6794 -1.5618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6794 -0.7525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3804 -0.3476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3804 -2.7374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0213 -0.3478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7358 -0.7604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4502 -0.3478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4502 0.4771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7358 0.8896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0213 0.4771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2907 -0.2862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7824 -2.7743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2088 0.8156 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7358 1.7145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7626 0.9074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0792 0.5128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2960 1.2716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0792 2.0351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7626 2.4297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5458 1.6708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3428 2.2725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1397 2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7935 0.9815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3860 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9644 0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7753 0.9338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5911 0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0129 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2019 1.2008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6028 1.2226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6197 1.6953 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6215 1.0812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1483 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1483 2.7743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9255 1.4281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1483 0.7311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9255 0.2824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
16 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
18 23 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
32 20 1 0 0 0 0
40 41 2 0 0 0 0
40 42 1 0 0 0 0
27 40 1 0 0 0 0
43 44 1 0 0 0 0
34 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 40 41 42
M SBL 1 1 46
M SMT 1 ^COOH
M SBV 1 46 0.6026 -0.2060
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 CH2OH
M SBV 2 48 -0.5572 -0.0290
S SKP 5
ID FL3FACGS0037
FORMULA C27H28O17
EXACTMASS 624.1326494699999
AVERAGEMASS 624.50102
SMILES O(c13)C(c(c4)cc(O)c(OC(O5)C(C(C(O)C(CO)5)O)O)c4)=CC(=O)c(c(cc(c3)OC(O2)C(C(C(O)C2C(O)=O)O)O)O)1
M END