Mol:FL3FACGS0064
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.3789 1.5401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3789 1.5401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3789 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3789 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0722 0.7176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0722 0.7176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5232 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5232 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5232 1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5232 1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0722 1.7592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0722 1.7592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9743 0.7176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9743 0.7176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4254 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4254 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4254 1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4254 1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9743 1.7592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9743 1.7592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9743 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9743 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8763 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8763 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3360 1.4937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3360 1.4937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7957 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7957 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7957 2.2900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7957 2.2900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3360 2.5554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3360 2.5554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8763 2.2900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8763 2.2900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0722 0.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0722 0.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8401 1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8401 1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2553 2.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2553 2.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1800 1.5373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1800 1.5373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3643 1.5037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3643 1.5037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8454 1.7815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8454 1.7815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3265 1.5037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3265 1.5037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3265 0.9482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3265 0.9482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8454 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8454 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3643 0.9482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3643 0.9482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8454 2.2308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8454 2.2308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7204 1.7312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7204 1.7312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6900 0.7383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6900 0.7383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8454 0.0683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8454 0.0683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3041 -0.1965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3041 -0.1965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3041 -0.8547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3041 -0.8547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8365 -1.1620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8365 -1.1620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8365 -1.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8365 -1.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3041 -2.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3041 -2.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7718 -1.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7718 -1.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7718 -1.1620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7718 -1.1620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2553 -0.9202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2553 -0.9202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2054 -1.9898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2054 -1.9898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3041 -2.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3041 -2.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4003 -2.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4003 -2.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0107 -1.9232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0107 -1.9232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 1 1 0 0 0 0 | + | 19 1 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 14 21 1 0 0 0 0 | + | 14 21 1 0 0 0 0 |
| − | 19 22 1 0 0 0 0 | + | 19 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 23 28 1 1 0 0 0 | + | 23 28 1 1 0 0 0 |
| − | 24 29 1 6 0 0 0 | + | 24 29 1 6 0 0 0 |
| − | 25 30 1 6 0 0 0 | + | 25 30 1 6 0 0 0 |
| − | 26 31 1 6 0 0 0 | + | 26 31 1 6 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 33 32 1 6 0 0 0 | + | 33 32 1 6 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 34 39 1 1 0 0 0 | + | 34 39 1 1 0 0 0 |
| − | 35 40 1 6 0 0 0 | + | 35 40 1 6 0 0 0 |
| − | 36 41 1 6 0 0 0 | + | 36 41 1 6 0 0 0 |
| − | 37 42 1 6 0 0 0 | + | 37 42 1 6 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACGS0064 | + | ID FL3FACGS0064 |
| − | KNApSAcK_ID C00004501 | + | KNApSAcK_ID C00004501 |
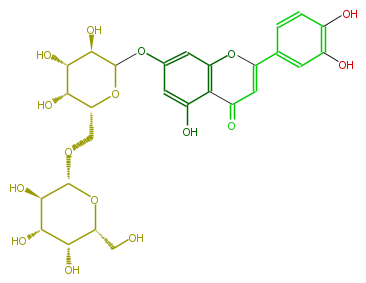
| − | NAME Luteolin 7-galactosyl-(1->6)-galactoside;2-(3,4-Dihydroxyphenyl)-7-[(6-O-beta-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-5-hydroxy-4H-1-benzopyran-4-one | + | NAME Luteolin 7-galactosyl-(1->6)-galactoside;2-(3,4-Dihydroxyphenyl)-7-[(6-O-beta-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-5-hydroxy-4H-1-benzopyran-4-one |
| − | CAS_RN 142561-45-3 | + | CAS_RN 142561-45-3 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(c3)(c(c(O)cc(OC(C(O)4)OC(COC(C5O)OC(C(C(O)5)O)CO)C(O)C4O)3)2)OC(=CC(=O)2)c(c1)cc(c(c1)O)O | + | SMILES c(c3)(c(c(O)cc(OC(C(O)4)OC(COC(C5O)OC(C(C(O)5)O)CO)C(O)C4O)3)2)OC(=CC(=O)2)c(c1)cc(c(c1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.3789 1.5401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3789 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0722 0.7176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5232 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5232 1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0722 1.7592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9743 0.7176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4254 0.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4254 1.4988 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9743 1.7592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9743 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8763 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3360 1.4937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7957 1.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7957 2.2900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3360 2.5554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8763 2.2900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0722 0.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8401 1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2553 2.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1800 1.5373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3643 1.5037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8454 1.7815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3265 1.5037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3265 0.9482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8454 0.6705 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3643 0.9482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8454 2.2308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7204 1.7312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6900 0.7383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8454 0.0683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3041 -0.1965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3041 -0.8547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8365 -1.1620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8365 -1.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3041 -2.0841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7718 -1.7767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7718 -1.1620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2553 -0.9202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2054 -1.9898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3041 -2.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4003 -2.1481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0107 -1.9232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 1 1 0 0 0 0
15 20 1 0 0 0 0
14 21 1 0 0 0 0
19 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
23 28 1 1 0 0 0
24 29 1 6 0 0 0
25 30 1 6 0 0 0
26 31 1 6 0 0 0
31 32 1 0 0 0 0
33 32 1 6 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
34 39 1 1 0 0 0
35 40 1 6 0 0 0
36 41 1 6 0 0 0
37 42 1 6 0 0 0
42 43 1 0 0 0 0
S SKP 8
ID FL3FACGS0064
KNApSAcK_ID C00004501
NAME Luteolin 7-galactosyl-(1->6)-galactoside;2-(3,4-Dihydroxyphenyl)-7-[(6-O-beta-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-5-hydroxy-4H-1-benzopyran-4-one
CAS_RN 142561-45-3
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(c3)(c(c(O)cc(OC(C(O)4)OC(COC(C5O)OC(C(C(O)5)O)CO)C(O)C4O)3)2)OC(=CC(=O)2)c(c1)cc(c(c1)O)O
M END