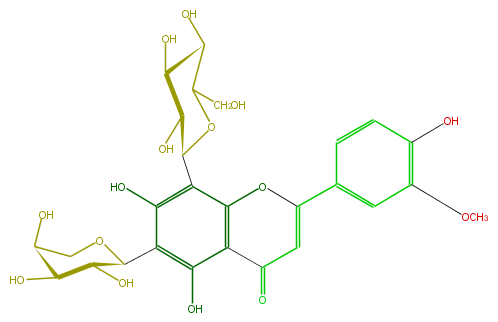
Mol:FL3FADCS0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3138 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3138 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3138 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3138 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5993 -2.1688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5993 -2.1688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8849 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8849 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8849 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8849 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5993 -0.5189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5993 -0.5189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1704 -2.1688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1704 -2.1688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5441 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5441 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5441 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5441 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1704 -0.5189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1704 -0.5189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1704 -2.8120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1704 -2.8120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0280 -0.5190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0280 -0.5190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3378 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3378 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0906 -0.9271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0906 -0.9271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8435 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8435 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8435 0.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8435 0.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0906 0.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0906 0.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3378 0.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3378 0.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5960 0.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5960 0.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3519 2.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3519 2.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1296 1.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1296 1.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7997 0.9169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7997 0.9169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7908 0.0981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7908 0.0981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1958 0.6932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1958 0.6932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5875 1.4354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5875 1.4354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7207 2.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7207 2.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1296 2.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1296 2.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1826 0.2536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1826 0.2536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5993 -2.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5993 -2.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8015 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8015 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3248 -2.3733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3248 -2.3733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6383 -2.1063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6383 -2.1063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9759 -2.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9759 -2.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4573 -1.6177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4573 -1.6177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0578 -1.9347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0578 -1.9347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6235 -1.0795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6235 -1.0795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0679 -2.4094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0679 -2.4094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9985 -2.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9985 -2.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9557 -1.1347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9557 -1.1347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0679 -1.7770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0679 -1.7770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0828 1.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0828 1.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4406 2.2562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4406 2.2562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 9 1 0 0 0 0 | + | 13 9 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 6 23 1 0 0 0 0 | + | 6 23 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 15 39 1 0 0 0 0 | + | 15 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -1.1122 0.6422 | + | M SBV 1 44 -1.1122 0.6422 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 -0.5047 0.2914 | + | M SBV 2 46 -0.5047 0.2914 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FADCS0007 | + | ID FL3FADCS0007 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES C(C5O)OC(C(C5O)O)c(c1O)c(c(C(=O)4)c(OC(=C4)c(c3)cc(OC)c(O)c3)c1C(O2)C(C(C(O)C2CO)O)O)O | + | SMILES C(C5O)OC(C(C5O)O)c(c1O)c(c(C(=O)4)c(OC(=C4)c(c3)cc(OC)c(O)c3)c1C(O2)C(C(C(O)C2CO)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.3138 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3138 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5993 -2.1688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8849 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8849 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5993 -0.5189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1704 -2.1688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5441 -1.7563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5441 -0.9314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1704 -0.5189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1704 -2.8120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0280 -0.5190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3378 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0906 -0.9271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8435 -0.4925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8435 0.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0906 0.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3378 0.3768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5960 0.8113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3519 2.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1296 1.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7997 0.9169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7908 0.0981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1958 0.6932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5875 1.4354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7207 2.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1296 2.4886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1826 0.2536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5993 -2.9935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8015 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3248 -2.3733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6383 -2.1063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9759 -2.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4573 -1.6177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0578 -1.9347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6235 -1.0795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0679 -2.4094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9985 -2.4757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9557 -1.1347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0679 -1.7770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0828 1.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4406 2.2562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
13 9 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
6 23 1 0 0 0 0
3 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 2 1 0 0 0 0
39 40 1 0 0 0 0
15 39 1 0 0 0 0
41 42 1 0 0 0 0
25 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -1.1122 0.6422
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 -0.5047 0.2914
S SKP 5
ID FL3FADCS0007
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(C5O)OC(C(C5O)O)c(c1O)c(c(C(=O)4)c(OC(=C4)c(c3)cc(OC)c(O)c3)c1C(O2)C(C(C(O)C2CO)O)O)O
M END