Mol:FL3FAECS0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4920 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4920 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4920 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4920 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9357 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9357 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3794 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3794 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3794 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3794 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9357 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9357 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8231 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8231 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2668 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2668 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2668 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2668 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8231 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8231 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8231 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8231 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0481 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0481 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7110 1.7972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7110 1.7972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3165 1.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3165 1.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0596 0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0596 0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0528 0.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0528 0.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5894 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5894 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8945 1.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8945 1.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1790 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1790 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3514 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3514 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6951 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6951 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9357 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9357 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4130 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4130 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9992 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9992 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5854 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5854 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5854 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5854 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9992 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9992 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4130 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4130 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9992 1.1139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9992 1.1139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1211 -1.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1211 -1.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8202 -2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8202 -2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3987 -1.8433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3987 -1.8433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9807 -2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9807 -2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2816 -1.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2816 -1.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7031 -1.6529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7031 -1.6529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5624 -1.6374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5624 -1.6374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9413 -1.2404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9413 -1.2404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7156 -1.7383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7156 -1.7383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3192 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3192 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6047 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6047 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3337 -1.9820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3337 -1.9820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0481 -2.3945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0481 -2.3945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1713 0.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1713 0.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8858 0.0247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8858 0.0247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 9 23 1 0 0 0 0 | + | 9 23 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 31 8 1 0 0 0 0 | + | 31 8 1 0 0 0 0 |
| − | 18 39 1 0 0 0 0 | + | 18 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 43 -6.9658 5.9300 | + | M SBV 1 43 -6.9658 5.9300 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 45 -7.1881 5.6246 | + | M SBV 2 45 -7.1881 5.6246 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 47 -6.9552 5.9361 | + | M SBV 3 47 -6.9552 5.9361 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAECS0006 | + | ID FL3FAECS0006 |
| − | KNApSAcK_ID C00006287 | + | KNApSAcK_ID C00006287 |
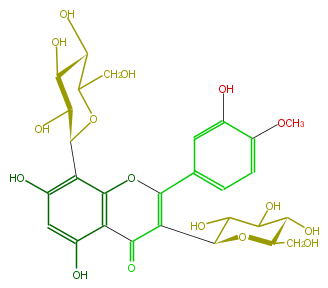
| − | NAME 3,8-Di-C-glucopyranosyldiosmetin | + | NAME 3,8-Di-C-glucopyranosyldiosmetin |
| − | CAS_RN 97218-32-1 | + | CAS_RN 97218-32-1 |
| − | FORMULA C28H32O16 | + | FORMULA C28H32O16 |
| − | EXACTMASS 624.1690349759999 | + | EXACTMASS 624.1690349759999 |
| − | AVERAGEMASS 624.54408 | + | AVERAGEMASS 624.54408 |
| − | SMILES O(C1c(c5O)c(c(c(c5)O)2)OC(c(c4)cc(O)c(c4)OC)=C(C(O3)C(O)C(O)C(C(CO)3)O)C2=O)C(CO)C(C(C1O)O)O | + | SMILES O(C1c(c5O)c(c(c(c5)O)2)OC(c(c4)cc(O)c(c4)OC)=C(C(O3)C(O)C(O)C(C(CO)3)O)C2=O)C(CO)C(C(C1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-2.4920 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4920 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9357 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3794 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3794 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9357 -0.6810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8231 -1.9657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2668 -1.6445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2668 -1.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8231 -0.6810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8231 -2.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0481 -0.6811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7110 1.7972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3165 1.3385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0596 0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0528 0.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5894 0.5036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8945 1.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1790 2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3514 2.0536 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6951 0.2993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9357 -2.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4130 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9992 -0.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5854 -0.5780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5854 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9992 0.4374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4130 0.0989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9992 1.1139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1211 -1.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8202 -2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3987 -1.8433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9807 -2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2816 -1.4877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7031 -1.6529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5624 -1.6374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9413 -1.2404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7156 -1.7383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3192 1.4137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6047 1.0012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3337 -1.9820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0481 -2.3945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1713 0.4372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8858 0.0247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
9 23 1 0 0 0 0
27 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
31 8 1 0 0 0 0
18 39 1 0 0 0 0
39 40 1 0 0 0 0
33 41 1 0 0 0 0
41 42 1 0 0 0 0
26 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 CH2OH
M SBV 1 43 -6.9658 5.9300
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 CH2OH
M SBV 2 45 -7.1881 5.6246
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 47
M SMT 3 OCH3
M SBV 3 47 -6.9552 5.9361
S SKP 8
ID FL3FAECS0006
KNApSAcK_ID C00006287
NAME 3,8-Di-C-glucopyranosyldiosmetin
CAS_RN 97218-32-1
FORMULA C28H32O16
EXACTMASS 624.1690349759999
AVERAGEMASS 624.54408
SMILES O(C1c(c5O)c(c(c(c5)O)2)OC(c(c4)cc(O)c(c4)OC)=C(C(O3)C(O)C(O)C(C(CO)3)O)C2=O)C(CO)C(C(C1O)O)O
M END