Mol:FL5FAAGL0022
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.9632 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9632 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9632 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9632 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2487 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2487 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5342 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5342 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5342 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5342 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2487 2.1296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2487 2.1296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8198 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8198 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1053 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1053 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1053 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1053 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8198 2.1296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8198 2.1296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8198 -0.1637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8198 -0.1637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6089 2.1293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6089 2.1293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3371 1.7089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3371 1.7089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0652 2.1293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0652 2.1293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0652 2.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0652 2.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3371 3.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3371 3.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6089 2.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6089 2.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2487 -0.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2487 -0.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6774 2.1293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6774 2.1293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7932 3.3905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7932 3.3905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4010 0.3925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4010 0.3925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4688 -1.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4688 -1.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2757 -0.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2757 -0.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3718 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3718 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7194 0.3384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7194 0.3384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9125 -0.1275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9125 -0.1275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8162 -0.3538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8162 -0.3538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9872 -0.1578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9872 -0.1578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6774 -1.0444 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6774 -1.0444 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0772 0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0772 0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2647 -1.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2647 -1.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4347 -1.9978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4347 -1.9978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7109 -2.0147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7109 -2.0147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2317 -2.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2317 -2.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9814 -2.4104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9814 -2.4104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7629 -2.4190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7629 -2.4190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1792 -1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1792 -1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4133 -2.1518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4133 -2.1518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0842 -2.4109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0842 -2.4109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6072 -2.6933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6072 -2.6933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6187 -2.8031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6187 -2.8031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6141 -2.2622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6141 -2.2622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2207 -3.0293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2207 -3.0293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4316 -2.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4316 -2.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6635 -3.0166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6635 -3.0166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1442 -2.3813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1442 -2.3813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9462 -2.5191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9462 -2.5191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2398 -2.5034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2398 -2.5034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0219 -2.9641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0219 -2.9641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8641 -3.3906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8641 -3.3906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4790 -1.8464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4790 -1.8464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6644 -1.3520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6644 -1.3520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8027 -1.6397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8027 -1.6397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4504 -1.3584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4504 -1.3584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 25 30 1 0 0 0 0 | + | 25 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 36 32 1 0 0 0 0 | + | 36 32 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 39 1 0 0 0 0 | + | 45 39 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 47 51 1 0 0 0 0 | + | 47 51 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 38 53 1 0 0 0 0 | + | 38 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 57 0.5327 -0.6727 | + | M SBV 1 57 0.5327 -0.6727 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 59 | + | M SBL 2 1 59 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 59 0.6106 -0.5121 | + | M SBV 2 59 0.6106 -0.5121 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0022 | + | ID FL5FAAGL0022 |
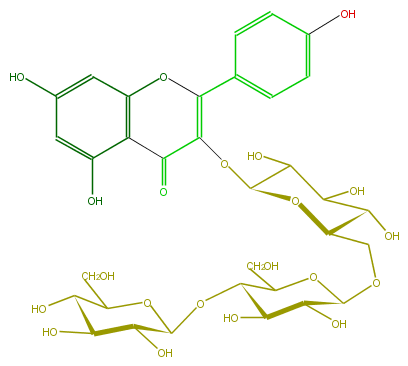
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES C(OC(C(O)6)OC(CO)C(O)C6O)(C1CO)C(O)C(C(OCC(C(O)5)OC(C(O)C5O)OC(C3=O)=C(Oc(c4)c3c(O)cc4O)c(c2)ccc(O)c2)O1)O | + | SMILES C(OC(C(O)6)OC(CO)C(O)C6O)(C1CO)C(O)C(C(OCC(C(O)5)OC(C(O)C5O)OC(C3=O)=C(Oc(c4)c3c(O)cc4O)c(c2)ccc(O)c2)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.9632 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9632 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2487 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5342 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5342 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2487 2.1296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8198 0.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1053 0.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1053 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8198 2.1296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8198 -0.1637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6089 2.1293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3371 1.7089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0652 2.1293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0652 2.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3371 3.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6089 2.9701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2487 -0.3452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6774 2.1293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7932 3.3905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4010 0.3925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4688 -1.0260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2757 -0.5601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3718 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7194 0.3384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9125 -0.1275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8162 -0.3538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9872 -0.1578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6774 -1.0444 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0772 0.5481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2647 -1.2393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4347 -1.9978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7109 -2.0147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2317 -2.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9814 -2.4104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7629 -2.4190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1792 -1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4133 -2.1518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0842 -2.4109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6072 -2.6933 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6187 -2.8031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6141 -2.2622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2207 -3.0293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4316 -2.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6635 -3.0166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1442 -2.3813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9462 -2.5191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2398 -2.5034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0219 -2.9641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8641 -3.3906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4790 -1.8464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6644 -1.3520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8027 -1.6397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4504 -1.3584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
26 21 1 0 0 0 0
25 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 41 1 0 0 0 0
36 32 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 39 1 0 0 0 0
51 52 1 0 0 0 0
47 51 1 0 0 0 0
53 54 1 0 0 0 0
38 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 ^ CH2OH
M SBV 1 57 0.5327 -0.6727
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 59
M SMT 2 ^ CH2OH
M SBV 2 59 0.6106 -0.5121
S SKP 5
ID FL5FAAGL0022
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES C(OC(C(O)6)OC(CO)C(O)C6O)(C1CO)C(O)C(C(OCC(C(O)5)OC(C(O)C5O)OC(C3=O)=C(Oc(c4)c3c(O)cc4O)c(c2)ccc(O)c2)O1)O
M END