Mol:FL5FAAGLS001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 36 39 0 0 0 0 0 0 0 0999 V2000 | + | 36 39 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.1033 -1.9163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1033 -1.9163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6113 -2.3292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6113 -2.3292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0077 -2.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0077 -2.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8962 -1.4768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8962 -1.4768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3882 -1.0640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3882 -1.0640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9918 -1.2837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9918 -1.2837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2925 -1.2570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2925 -1.2570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1810 -0.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1810 -0.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6730 -0.2117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6730 -0.2117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2767 -0.4314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2767 -0.4314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0912 -1.5790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0912 -1.5790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5635 0.6108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5635 0.6108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0516 0.8347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0516 0.8347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1654 1.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1654 1.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3362 1.9003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3362 1.9003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9514 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9514 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0651 1.0316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0651 1.0316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8244 -2.1787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8244 -2.1787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3633 2.4887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3633 2.4887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5766 -2.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5766 -2.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1578 -0.3381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1578 -0.3381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7579 -1.7917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7579 -1.7917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5519 -1.3498 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5519 -1.3498 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9211 -1.2509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9211 -1.2509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3686 -0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3686 -0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0943 -1.5631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0943 -1.5631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9883 -0.1300 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.9883 -0.1300 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.3900 -0.6602 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.3900 -0.6602 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.9684 -0.4353 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.9684 -0.4353 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.5266 -0.4292 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.5266 -0.4292 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1210 -0.0235 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.1210 -0.0235 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.6149 -0.2906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6149 -0.2906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7638 -0.6907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7638 -0.6907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5266 -0.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5266 -0.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3517 0.2185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3517 0.2185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0700 1.1780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0700 1.1780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 20 3 1 0 0 0 0 | + | 20 3 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 23 26 2 0 0 0 0 | + | 23 26 2 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 28 33 1 0 0 0 0 | + | 28 33 1 0 0 0 0 |
| − | 30 34 1 0 0 0 0 | + | 30 34 1 0 0 0 0 |
| − | 27 21 1 0 0 0 0 | + | 27 21 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 35 36 | + | M SAL 1 2 35 36 |
| − | M SBL 1 1 38 | + | M SBL 1 1 38 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 38 2.4547 -0.0447 | + | M SVB 1 38 2.4547 -0.0447 |
| − | S SKP 8 | + | S SKP 8 |
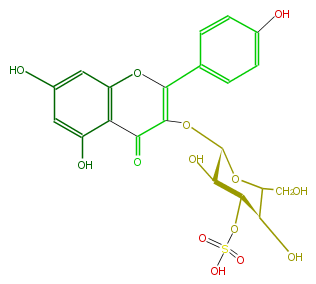
| − | ID FL5FAAGLS001 | + | ID FL5FAAGLS001 |
| − | KNApSAcK_ID C00006063 | + | KNApSAcK_ID C00006063 |
| − | NAME Kaempferol 3-(3''-sulfatoglucoside) | + | NAME Kaempferol 3-(3''-sulfatoglucoside) |
| − | CAS_RN 85290-33-1 | + | CAS_RN 85290-33-1 |
| − | FORMULA C21H20O14S | + | FORMULA C21H20O14S |
| − | EXACTMASS 528.057376038 | + | EXACTMASS 528.057376038 |
| − | AVERAGEMASS 528.4411 | + | AVERAGEMASS 528.4411 |
| − | SMILES OCC([C@@H](O)1)O[C@H](OC(C(=O)3)=C(Oc(c4)c3c(cc(O)4)O)c(c2)ccc(O)c2)[C@H](O)[C@@H]1OS(O)(=O)=O | + | SMILES OCC([C@@H](O)1)O[C@H](OC(C(=O)3)=C(Oc(c4)c3c(cc(O)4)O)c(c2)ccc(O)c2)[C@H](O)[C@@H]1OS(O)(=O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
36 39 0 0 0 0 0 0 0 0999 V2000
-2.1033 -1.9163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6113 -2.3292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0077 -2.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8962 -1.4768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3882 -1.0640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9918 -1.2837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2925 -1.2570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1810 -0.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6730 -0.2117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2767 -0.4314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0912 -1.5790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5635 0.6108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0516 0.8347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1654 1.4795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3362 1.9003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9514 1.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0651 1.0316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8244 -2.1787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3633 2.4887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5766 -2.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1578 -0.3381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7579 -1.7917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5519 -1.3498 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
2.9211 -1.2509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3686 -0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0943 -1.5631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9883 -0.1300 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.3900 -0.6602 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.9684 -0.4353 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.5266 -0.4292 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1210 -0.0235 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.6149 -0.2906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7638 -0.6907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5266 -0.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3517 0.2185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0700 1.1780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
4 3 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
20 3 1 0 0 0 0
22 23 1 0 0 0 0
23 24 2 0 0 0 0
23 25 1 0 0 0 0
23 26 2 0 0 0 0
8 21 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
28 33 1 0 0 0 0
30 34 1 0 0 0 0
27 21 1 0 0 0 0
25 29 1 0 0 0 0
31 35 1 0 0 0 0
35 36 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 35 36
M SBL 1 1 38
M SMT 1 CH2OH
M SVB 1 38 2.4547 -0.0447
S SKP 8
ID FL5FAAGLS001
KNApSAcK_ID C00006063
NAME Kaempferol 3-(3''-sulfatoglucoside)
CAS_RN 85290-33-1
FORMULA C21H20O14S
EXACTMASS 528.057376038
AVERAGEMASS 528.4411
SMILES OCC([C@@H](O)1)O[C@H](OC(C(=O)3)=C(Oc(c4)c3c(cc(O)4)O)c(c2)ccc(O)c2)[C@H](O)[C@@H]1OS(O)(=O)=O
M END