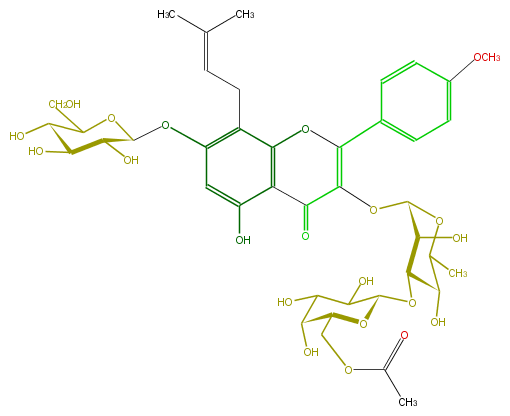
Mol:FL5FABGI0022
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 62 67 0 0 0 0 0 0 0 0999 V2000 | + | 62 67 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3888 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3888 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3888 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3888 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6741 0.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6741 0.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0407 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0407 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0407 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0407 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6741 1.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6741 1.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7555 0.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7555 0.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4702 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4702 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4702 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4702 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7555 1.6863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7555 1.6863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7555 -0.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7555 -0.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3703 1.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3703 1.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0988 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0988 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8271 1.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8271 1.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8271 2.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8271 2.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0988 3.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0988 3.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3703 2.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3703 2.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2426 1.7666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2426 1.7666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6741 -0.6873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6741 -0.6873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2204 -0.0513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2204 -0.0513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9370 -1.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9370 -1.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6249 -1.8136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6249 -1.8136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4067 -1.0499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4067 -1.0499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6249 -0.2816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6249 -0.2816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9370 0.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9370 0.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1553 -0.6481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1553 -0.6481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9988 -0.6481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9988 -0.6481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0396 -2.0414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0396 -2.0414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5245 -2.4529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5245 -2.4529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9017 -1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9017 -1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7668 1.7913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7668 1.7913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2899 1.1617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2899 1.1617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6031 1.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6031 1.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9404 1.4359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9404 1.4359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4219 1.9176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4219 1.9176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0227 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0227 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3637 1.5324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3637 1.5324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9494 1.2066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9494 1.2066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0624 1.0129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0624 1.0129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6741 2.5113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6741 2.5113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3886 2.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3886 2.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3886 3.7489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3886 3.7489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6741 4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6741 4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1031 4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1031 4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9862 -2.4962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9862 -2.4962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3302 -1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3302 -1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6687 -2.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6687 -2.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0034 -1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0034 -1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6594 -2.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6594 -2.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3208 -2.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3208 -2.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7912 -3.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7912 -3.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3913 -2.0271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3913 -2.0271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0853 -2.7376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0853 -2.7376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9713 -1.5789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9713 -1.5789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6704 -3.4846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6704 -3.4846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4356 -3.4846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4356 -3.4846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8676 -2.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8676 -2.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8264 -4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8264 -4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4457 3.2408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4457 3.2408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3637 2.7108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3637 2.7108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6984 2.2148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6984 2.2148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6745 1.9403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6745 1.9403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 19 3 1 0 0 0 0 | + | 19 3 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 18 1 0 0 0 0 | + | 34 18 1 0 0 0 0 |
| − | 6 40 1 0 0 0 0 | + | 6 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 1 0 0 0 | + | 49 50 1 1 0 0 0 |
| − | 50 45 1 1 0 0 0 | + | 50 45 1 1 0 0 0 |
| − | 49 51 1 0 0 0 0 | + | 49 51 1 0 0 0 0 |
| − | 48 52 1 0 0 0 0 | + | 48 52 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 47 54 1 0 0 0 0 | + | 47 54 1 0 0 0 0 |
| − | 46 28 1 0 0 0 0 | + | 46 28 1 0 0 0 0 |
| − | 53 55 1 0 0 0 0 | + | 53 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 56 57 2 0 0 0 0 | + | 56 57 2 0 0 0 0 |
| − | 56 58 1 0 0 0 0 | + | 56 58 1 0 0 0 0 |
| − | 59 60 1 0 0 0 0 | + | 59 60 1 0 0 0 0 |
| − | 15 59 1 0 0 0 0 | + | 15 59 1 0 0 0 0 |
| − | 61 62 1 0 0 0 0 | + | 61 62 1 0 0 0 0 |
| − | 36 61 1 0 0 0 0 | + | 36 61 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 59 60 | + | M SAL 1 2 59 60 |
| − | M SBL 1 1 65 | + | M SBL 1 1 65 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 65 -0.6185 -0.5545 | + | M SBV 1 65 -0.6185 -0.5545 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 61 62 | + | M SAL 2 2 61 62 |
| − | M SBL 2 1 67 | + | M SBL 2 1 67 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 67 0.6756 -0.6143 | + | M SBV 2 67 0.6756 -0.6143 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FABGI0022 | + | ID FL5FABGI0022 |
| − | FORMULA C41H52O21 | + | FORMULA C41H52O21 |
| − | EXACTMASS 880.300108726 | + | EXACTMASS 880.300108726 |
| − | AVERAGEMASS 880.83898 | + | AVERAGEMASS 880.83898 |
| − | SMILES C(C1O)(O)C(O)C(Oc(c2)c(CC=C(C)C)c(O5)c(C(=O)C(=C(c(c6)ccc(c6)OC)5)OC(C(O)3)OC(C(O)C3OC(C4O)OC(COC(C)=O)C(O)C4O)C)c(O)2)OC1CO | + | SMILES C(C1O)(O)C(O)C(Oc(c2)c(CC=C(C)C)c(O5)c(C(=O)C(=C(c(c6)ccc(c6)OC)5)OC(C(O)3)OC(C(O)C3OC(C4O)OC(COC(C)=O)C(O)C4O)C)c(O)2)OC1CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
62 67 0 0 0 0 0 0 0 0999 V2000
-1.3888 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3888 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6741 0.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0407 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0407 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6741 1.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7555 0.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4702 0.4483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4702 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7555 1.6863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7555 -0.6078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3703 1.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0988 1.4246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8271 1.8451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8271 2.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0988 3.1070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3703 2.6863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2426 1.7666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6741 -0.6873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2204 -0.0513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9370 -1.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6249 -1.8136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4067 -1.0499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6249 -0.2816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9370 0.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1553 -0.6481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9988 -0.6481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0396 -2.0414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5245 -2.4529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9017 -1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7668 1.7913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2899 1.1617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6031 1.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9404 1.4359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4219 1.9176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0227 1.6005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3637 1.5324 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9494 1.2066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0624 1.0129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6741 2.5113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3886 2.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3886 3.7489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6741 4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1031 4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9862 -2.4962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3302 -1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6687 -2.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0034 -1.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6594 -2.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3208 -2.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7912 -3.0935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3913 -2.0271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0853 -2.7376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9713 -1.5789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6704 -3.4846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4356 -3.4846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8676 -2.7363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8264 -4.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4457 3.2408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3637 2.7108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6984 2.2148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6745 1.9403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
4 3 1 0 0 0 0
1 18 1 0 0 0 0
19 3 1 0 0 0 0
8 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
25 20 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 18 1 0 0 0 0
6 40 1 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 43 1 0 0 0 0
42 44 1 0 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 50 1 1 0 0 0
50 45 1 1 0 0 0
49 51 1 0 0 0 0
48 52 1 0 0 0 0
50 53 1 0 0 0 0
47 54 1 0 0 0 0
46 28 1 0 0 0 0
53 55 1 0 0 0 0
55 56 1 0 0 0 0
56 57 2 0 0 0 0
56 58 1 0 0 0 0
59 60 1 0 0 0 0
15 59 1 0 0 0 0
61 62 1 0 0 0 0
36 61 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 59 60
M SBL 1 1 65
M SMT 1 OCH3
M SBV 1 65 -0.6185 -0.5545
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 61 62
M SBL 2 1 67
M SMT 2 ^CH2OH
M SBV 2 67 0.6756 -0.6143
S SKP 5
ID FL5FABGI0022
FORMULA C41H52O21
EXACTMASS 880.300108726
AVERAGEMASS 880.83898
SMILES C(C1O)(O)C(O)C(Oc(c2)c(CC=C(C)C)c(O5)c(C(=O)C(=C(c(c6)ccc(c6)OC)5)OC(C(O)3)OC(C(O)C3OC(C4O)OC(COC(C)=O)C(O)C4O)C)c(O)2)OC1CO
M END