Mol:FL5FACGA0037
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 59 64 0 0 0 0 0 0 0 0999 V2000 | + | 59 64 0 0 0 0 0 0 0 0999 V2000 |
| − | -5.2468 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2468 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2468 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2468 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5322 -0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5322 -0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8176 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8176 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8176 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8176 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5322 0.9479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5322 0.9479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1031 -0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1031 -0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3886 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3886 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3886 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3886 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1031 0.9479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1031 0.9479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1031 -1.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1031 -1.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6744 0.9478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6744 0.9478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9462 0.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9462 0.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2180 0.9478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2180 0.9478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2180 1.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2180 1.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9462 2.2092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9462 2.2092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6744 1.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6744 1.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9609 0.9478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9609 0.9478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5100 2.2091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5100 2.2091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7353 -0.8950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7353 -0.8950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5322 -1.5269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5322 -1.5269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2866 -2.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2866 -2.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9378 -1.5806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9378 -1.5806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7693 -2.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7693 -2.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9937 -3.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9937 -3.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2866 -3.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2866 -3.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5109 -2.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5109 -2.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9844 -3.2468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9844 -3.2468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5860 -3.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5860 -3.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2413 -1.2952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2413 -1.2952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3719 -1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3719 -1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1440 -1.6946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1440 -1.6946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8870 -1.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8870 -1.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6445 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6445 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0829 -0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0829 -0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4330 -1.2199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4330 -1.2199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4733 -1.6732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4733 -1.6732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5629 -1.8549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5629 -1.8549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1311 -1.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1311 -1.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0095 -0.7985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0095 -0.7985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4564 -0.3747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4564 -0.3747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2281 0.0707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2281 0.0707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8693 -0.2995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8693 -0.2995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2281 0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2281 0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8594 1.3253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8594 1.3253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8594 2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8594 2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4514 2.3947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4514 2.3947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4514 3.0783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4514 3.0783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8594 3.4200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8594 3.4200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2674 3.0783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2674 3.0783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2674 2.3947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2674 2.3947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8594 4.1036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8594 4.1036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9462 3.0499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9462 3.0499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0434 3.4200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0434 3.4200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9609 2.8902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9609 2.8902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8871 -4.0251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8871 -4.0251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3349 -4.9855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3349 -4.9855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8086 3.8730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8086 3.8730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1665 4.9855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1665 4.9855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 31 30 1 0 0 0 0 | + | 31 30 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 46 1 0 0 0 0 | + | 51 46 1 0 0 0 0 |
| − | 49 52 1 0 0 0 0 | + | 49 52 1 0 0 0 0 |
| − | 16 53 1 0 0 0 0 | + | 16 53 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 48 54 1 0 0 0 0 | + | 48 54 1 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | 25 56 1 0 0 0 0 | + | 25 56 1 0 0 0 0 |
| − | 58 59 1 0 0 0 0 | + | 58 59 1 0 0 0 0 |
| − | 50 58 1 0 0 0 0 | + | 50 58 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 60 | + | M SBL 1 1 60 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 60 -0.5920 -0.3418 | + | M SBV 1 60 -0.5920 -0.3418 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 56 57 | + | M SAL 2 2 56 57 |
| − | M SBL 2 1 62 | + | M SBL 2 1 62 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 62 -0.1066 0.7799 | + | M SBV 2 62 -0.1066 0.7799 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 58 59 | + | M SAL 3 2 58 59 |
| − | M SBL 3 1 64 | + | M SBL 3 1 64 |
| − | M SMT 3 ^ OCH3 | + | M SMT 3 ^ OCH3 |
| − | M SBV 3 64 0.4588 -0.7948 | + | M SBV 3 64 0.4588 -0.7948 |
| − | S SKP 5 | + | S SKP 5 |
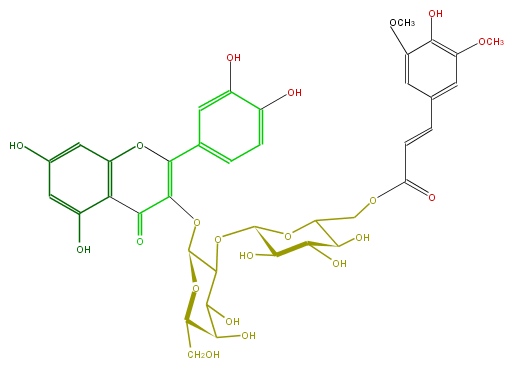
| − | ID FL5FACGA0037 | + | ID FL5FACGA0037 |
| − | FORMULA C38H40O21 | + | FORMULA C38H40O21 |
| − | EXACTMASS 832.206208342 | + | EXACTMASS 832.206208342 |
| − | AVERAGEMASS 832.7116 | + | AVERAGEMASS 832.7116 |
| − | SMILES c(c15)c(O)cc(c1C(=O)C(=C(c(c6)ccc(O)c(O)6)O5)OC(C(OC(C3O)OC(COC(C=Cc(c4)cc(c(c(OC)4)O)OC)=O)C(C(O)3)O)2)OC(C(O)C(O)2)CO)O | + | SMILES c(c15)c(O)cc(c1C(=O)C(=C(c(c6)ccc(O)c(O)6)O5)OC(C(OC(C3O)OC(COC(C=Cc(c4)cc(c(c(OC)4)O)OC)=O)C(C(O)3)O)2)OC(C(O)C(O)2)CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
59 64 0 0 0 0 0 0 0 0999 V2000
-5.2468 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2468 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5322 -0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8176 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8176 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5322 0.9479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1031 -0.7021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3886 -0.2896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3886 0.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1031 0.9479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1031 -1.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6744 0.9478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9462 0.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2180 0.9478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2180 1.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9462 2.2092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6744 1.7887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9609 0.9478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5100 2.2091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7353 -0.8950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5322 -1.5269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2866 -2.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9378 -1.5806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7693 -2.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9937 -3.2452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2866 -3.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5109 -2.8685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9844 -3.2468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5860 -3.5924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2413 -1.2952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3719 -1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1440 -1.6946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8870 -1.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6445 -1.4905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0829 -0.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4330 -1.2199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4733 -1.6732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5629 -1.8549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1311 -1.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0095 -0.7985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4564 -0.3747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2281 0.0707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8693 -0.2995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2281 0.9608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8594 1.3253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8594 2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4514 2.3947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4514 3.0783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8594 3.4200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2674 3.0783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2674 2.3947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8594 4.1036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9462 3.0499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0434 3.4200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9609 2.8902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8871 -4.0251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3349 -4.9855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8086 3.8730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1665 4.9855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
26 29 1 0 0 0 0
22 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
35 40 1 0 0 0 0
23 20 1 0 0 0 0
31 30 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 46 1 0 0 0 0
49 52 1 0 0 0 0
16 53 1 0 0 0 0
54 55 1 0 0 0 0
48 54 1 0 0 0 0
56 57 1 0 0 0 0
25 56 1 0 0 0 0
58 59 1 0 0 0 0
50 58 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 60
M SMT 1 OCH3
M SBV 1 60 -0.5920 -0.3418
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 56 57
M SBL 2 1 62
M SMT 2 CH2OH
M SBV 2 62 -0.1066 0.7799
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 58 59
M SBL 3 1 64
M SMT 3 ^ OCH3
M SBV 3 64 0.4588 -0.7948
S SKP 5
ID FL5FACGA0037
FORMULA C38H40O21
EXACTMASS 832.206208342
AVERAGEMASS 832.7116
SMILES c(c15)c(O)cc(c1C(=O)C(=C(c(c6)ccc(O)c(O)6)O5)OC(C(OC(C3O)OC(COC(C=Cc(c4)cc(c(c(OC)4)O)OC)=O)C(C(O)3)O)2)OC(C(O)C(O)2)CO)O
M END