Mol:FL5FACGS0076
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.1147 4.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1147 4.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1147 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1147 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8291 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8291 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5436 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5436 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5436 4.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5436 4.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8291 4.6703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8291 4.6703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4002 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4002 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3143 3.4328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3143 3.4328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0288 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0288 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0288 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0288 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3143 1.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3143 1.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4002 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4002 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7432 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7432 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4577 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4577 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4577 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4577 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7432 1.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7432 1.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3143 0.9578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3143 0.9578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1722 3.4328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1722 3.4328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1147 1.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1147 1.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7432 0.9578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7432 0.9578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1355 4.5995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1355 4.5995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1722 3.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1722 3.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1886 -0.0240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1886 -0.0240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6017 -0.2616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6017 -0.2616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0397 0.4378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0397 0.4378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7730 0.8165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7730 0.8165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9827 1.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9827 1.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5446 0.3547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5446 0.3547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0288 0.6737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0288 0.6737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5105 0.5466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5105 0.5466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2093 -0.7070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2093 -0.7070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0975 -0.3805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0975 -0.3805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3535 -0.1457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3535 -0.1457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5703 -2.1604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5703 -2.1604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2422 -2.3052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2422 -2.3052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5965 -1.5598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5965 -1.5598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2812 -1.0990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2812 -1.0990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4687 -0.9542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4687 -0.9542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1143 -1.6995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1143 -1.6995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4746 -1.4236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4746 -1.4236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9958 -1.6122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9958 -1.6122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0601 -2.2024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0601 -2.2024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4926 -2.8177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4926 -2.8177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1902 -1.8380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1902 -1.8380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4261 -3.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4261 -3.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3913 -4.1055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3913 -4.1055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7166 -3.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7166 -3.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3830 -2.8602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3830 -2.8602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5655 -2.7468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5655 -2.7468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2402 -3.5053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2402 -3.5053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3186 -3.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3186 -3.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8319 -3.4779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8319 -3.4779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3416 -4.6703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3416 -4.6703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0887 -4.3005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0887 -4.3005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1163 -3.8754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1163 -3.8754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 26 19 1 0 0 0 0 | + | 26 19 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 37 33 1 0 0 0 0 | + | 37 33 1 0 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 46 47 1 1 0 0 0 | + | 46 47 1 1 0 0 0 |
| − | 48 47 1 1 0 0 0 | + | 48 47 1 1 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 45 53 1 0 0 0 0 | + | 45 53 1 0 0 0 0 |
| − | 46 54 1 0 0 0 0 | + | 46 54 1 0 0 0 0 |
| − | 47 55 1 0 0 0 0 | + | 47 55 1 0 0 0 0 |
| − | 48 44 1 0 0 0 0 | + | 48 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0076 | + | ID FL5FACGS0076 |
| − | KNApSAcK_ID C00013848 | + | KNApSAcK_ID C00013848 |
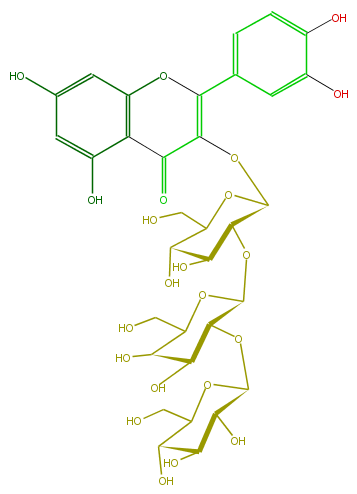
| − | NAME Quercetin 3-glucosyl-(1->2)-galactosyl-(1->2)-glucoside;3-[(O-beta-D-Glucopyranosyl-(1->2)-O-beta-D-galactopyranosyl-(1->2)-beta-D-glucopyranosyl)oxy]-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-1-benzopyran-4-one | + | NAME Quercetin 3-glucosyl-(1->2)-galactosyl-(1->2)-glucoside;3-[(O-beta-D-Glucopyranosyl-(1->2)-O-beta-D-galactopyranosyl-(1->2)-beta-D-glucopyranosyl)oxy]-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-1-benzopyran-4-one |
| − | CAS_RN 197250-97-8 | + | CAS_RN 197250-97-8 |
| − | FORMULA C33H40O22 | + | FORMULA C33H40O22 |
| − | EXACTMASS 788.201122964 | + | EXACTMASS 788.201122964 |
| − | AVERAGEMASS 788.6575 | + | AVERAGEMASS 788.6575 |
| − | SMILES O(C1OC(C(O)6)C(OC(C6O)CO)OC(=C(c(c5)ccc(O)c5O)3)C(=O)c(c4O)c(cc(O)c4)O3)C(CO)C(O)C(C(OC(O2)C(O)C(O)C(O)C2CO)1)O | + | SMILES O(C1OC(C(O)6)C(OC(C6O)CO)OC(=C(c(c5)ccc(O)c5O)3)C(=O)c(c4O)c(cc(O)c4)O3)C(CO)C(O)C(C(OC(O2)C(O)C(O)C(O)C2CO)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
1.1147 4.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1147 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8291 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5436 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5436 4.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8291 4.6703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4002 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3143 3.4328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0288 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0288 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3143 1.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4002 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7432 3.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4577 3.0203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4577 2.1953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7432 1.7828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3143 0.9578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1722 3.4328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1147 1.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7432 0.9578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1355 4.5995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1722 3.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1886 -0.0240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6017 -0.2616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0397 0.4378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7730 0.8165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9827 1.0542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5446 0.3547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0288 0.6737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5105 0.5466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2093 -0.7070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0975 -0.3805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3535 -0.1457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5703 -2.1604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2422 -2.3052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5965 -1.5598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2812 -1.0990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4687 -0.9542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1143 -1.6995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4746 -1.4236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9958 -1.6122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0601 -2.2024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4926 -2.8177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1902 -1.8380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4261 -3.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3913 -4.1055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7166 -3.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3830 -2.8602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5655 -2.7468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2402 -3.5053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3186 -3.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8319 -3.4779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3416 -4.6703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0887 -4.3005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1163 -3.8754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
26 19 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
35 43 1 0 0 0 0
36 44 1 0 0 0 0
37 33 1 0 0 0 0
45 46 1 1 0 0 0
46 47 1 1 0 0 0
48 47 1 1 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
50 45 1 0 0 0 0
50 51 1 0 0 0 0
51 52 1 0 0 0 0
45 53 1 0 0 0 0
46 54 1 0 0 0 0
47 55 1 0 0 0 0
48 44 1 0 0 0 0
S SKP 8
ID FL5FACGS0076
KNApSAcK_ID C00013848
NAME Quercetin 3-glucosyl-(1->2)-galactosyl-(1->2)-glucoside;3-[(O-beta-D-Glucopyranosyl-(1->2)-O-beta-D-galactopyranosyl-(1->2)-beta-D-glucopyranosyl)oxy]-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-1-benzopyran-4-one
CAS_RN 197250-97-8
FORMULA C33H40O22
EXACTMASS 788.201122964
AVERAGEMASS 788.6575
SMILES O(C1OC(C(O)6)C(OC(C6O)CO)OC(=C(c(c5)ccc(O)c5O)3)C(=O)c(c4O)c(cc(O)c4)O3)C(CO)C(O)C(C(OC(O2)C(O)C(O)C(O)C2CO)1)O
M END