Mol:FL5FADGA0021
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 52 0 0 0 0 0 0 0 0999 V2000 | + | 48 52 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.7043 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7043 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7043 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7043 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1480 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1480 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5917 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5917 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5917 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5917 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1480 2.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1480 2.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0354 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0354 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4791 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4791 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4791 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4791 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0354 2.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0354 2.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0354 0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0354 0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2214 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2214 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7883 1.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7883 1.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3553 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3553 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3553 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3553 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7883 3.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7883 3.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2214 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2214 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1480 0.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1480 0.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2214 3.3737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2214 3.3737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2199 2.2280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2199 2.2280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8917 1.3265 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.8917 1.3265 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.2934 0.7963 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2934 0.7963 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.8719 1.0212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.8719 1.0212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4301 1.0273 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4301 1.0273 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1276 1.4536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.1276 1.4536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.5184 1.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5184 1.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3356 1.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3356 1.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9142 0.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9142 0.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8242 0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8242 0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8404 0.8359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8404 0.8359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8957 -0.8373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.8957 -0.8373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.6257 -0.2583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.6257 -0.2583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.4173 -0.8621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4173 -0.8621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9189 -1.2676 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9189 -1.2676 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.1889 -1.8467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.1889 -1.8467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.3975 -1.2428 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.3975 -1.2428 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.1058 -0.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1058 -0.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7849 -1.5680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7849 -1.5680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1889 -2.2495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1889 -2.2495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2789 -2.1816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2789 -2.1816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4389 -2.5164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4389 -2.5164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9888 -2.1312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9888 -2.1312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9466 -1.6488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9466 -1.6488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4450 -2.5139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4450 -2.5139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0718 3.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0718 3.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5133 4.6744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5133 4.6744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3583 1.6956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3583 1.6956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1335 1.9777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1335 1.9777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 32 29 1 0 0 0 0 | + | 32 29 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 16 45 1 0 0 0 0 | + | 16 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 25 47 1 0 0 0 0 | + | 25 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 47 48 | + | M SAL 2 2 47 48 |
| − | M SBL 2 1 51 | + | M SBL 2 1 51 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 51 2.4613 1.4324 | + | M SVB 2 51 2.4613 1.4324 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 45 46 | + | M SAL 1 2 45 46 |
| − | M SBL 1 1 49 | + | M SBL 1 1 49 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 49 1.0718 3.7772 | + | M SVB 1 49 1.0718 3.7772 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGA0021 | + | ID FL5FADGA0021 |
| − | KNApSAcK_ID C00006012 | + | KNApSAcK_ID C00006012 |
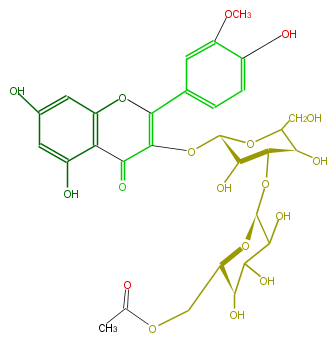
| − | NAME Isorhamnetin 3-(6'''-acetylglucosyl)(1->3)-galactoside;3-[[3-O-(6-O-Acetyl-beta-D-glucopyranosyl)-beta-D-galactopyranosyl]oxy]-5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4H-1-benzopyran-4-one | + | NAME Isorhamnetin 3-(6'''-acetylglucosyl)(1->3)-galactoside;3-[[3-O-(6-O-Acetyl-beta-D-glucopyranosyl)-beta-D-galactopyranosyl]oxy]-5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 139955-72-9 | + | CAS_RN 139955-72-9 |
| − | FORMULA C30H34O18 | + | FORMULA C30H34O18 |
| − | EXACTMASS 682.174514284 | + | EXACTMASS 682.174514284 |
| − | AVERAGEMASS 682.58016 | + | AVERAGEMASS 682.58016 |
| − | SMILES c(C(=C(O[C@@H](O4)[C@H](O)[C@H](O[C@H](O5)C(C(O)[C@@H](O)[C@@H](COC(C)=O)5)O)[C@@H](C4CO)O)3)Oc(c2)c(C(=O)3)c(cc(O)2)O)(c1)cc(c(O)c1)OC | + | SMILES c(C(=C(O[C@@H](O4)[C@H](O)[C@H](O[C@H](O5)C(C(O)[C@@H](O)[C@@H](COC(C)=O)5)O)[C@@H](C4CO)O)3)Oc(c2)c(C(=O)3)c(cc(O)2)O)(c1)cc(c(O)c1)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 52 0 0 0 0 0 0 0 0999 V2000
-2.7043 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7043 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1480 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5917 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5917 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1480 2.0954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0354 0.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4791 1.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4791 1.7742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0354 2.0954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0354 0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2214 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7883 1.8917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3553 2.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3553 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7883 3.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2214 2.8737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1480 0.1686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2214 3.3737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2199 2.2280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8917 1.3265 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.2934 0.7963 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.8719 1.0212 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4301 1.0273 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1276 1.4536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.5184 1.1658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3356 1.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9142 0.2971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8242 0.3753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8404 0.8359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8957 -0.8373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.6257 -0.2583 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.4173 -0.8621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9189 -1.2676 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.1889 -1.8467 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.3975 -1.2428 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.1058 -0.2601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7849 -1.5680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1889 -2.2495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2789 -2.1816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4389 -2.5164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9888 -2.1312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9466 -1.6488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4450 -2.5139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0718 3.7772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5133 4.6744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3583 1.6956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1335 1.9777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
27 8 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
34 40 1 0 0 0 0
32 29 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
16 45 1 0 0 0 0
45 46 1 0 0 0 0
25 47 1 0 0 0 0
47 48 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 47 48
M SBL 2 1 51
M SMT 2 CH2OH
M SVB 2 51 2.4613 1.4324
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 45 46
M SBL 1 1 49
M SMT 1 OCH3
M SVB 1 49 1.0718 3.7772
S SKP 8
ID FL5FADGA0021
KNApSAcK_ID C00006012
NAME Isorhamnetin 3-(6'''-acetylglucosyl)(1->3)-galactoside;3-[[3-O-(6-O-Acetyl-beta-D-glucopyranosyl)-beta-D-galactopyranosyl]oxy]-5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4H-1-benzopyran-4-one
CAS_RN 139955-72-9
FORMULA C30H34O18
EXACTMASS 682.174514284
AVERAGEMASS 682.58016
SMILES c(C(=C(O[C@@H](O4)[C@H](O)[C@H](O[C@H](O5)C(C(O)[C@@H](O)[C@@H](COC(C)=O)5)O)[C@@H](C4CO)O)3)Oc(c2)c(C(=O)3)c(cc(O)2)O)(c1)cc(c(O)c1)OC
M END