Mol:FL5FADGL0038
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.9580 -0.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9580 -0.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6866 -1.0953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6866 -1.0953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0467 -1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0467 -1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6782 -0.6251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6782 -0.6251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9497 -0.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9497 -0.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5896 0.0130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5896 0.0130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0383 -0.6811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0383 -0.6811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6699 -0.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6699 -0.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9413 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9413 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5813 0.4832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5813 0.4832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8266 -1.1349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8266 -1.1349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5730 0.9532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5730 0.9532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0792 0.8962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0792 0.8962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4547 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4547 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1780 2.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1780 2.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4742 2.0829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4742 2.0829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8497 1.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8497 1.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1501 -0.3798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1501 -0.3798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2099 0.4065 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.2099 0.4065 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.8223 -0.2649 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.8223 -0.2649 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.5678 -0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5678 -0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3177 -0.2649 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.3177 -0.2649 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7055 0.4065 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7055 0.4065 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.9600 0.1935 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.9600 0.1935 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.4829 0.2117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4829 0.2117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2288 0.6591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2288 0.6591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3197 0.2419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3197 0.2419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7516 2.8450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7516 2.8450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7753 -1.7331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7753 -1.7331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5956 -0.5915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5956 -0.5915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9467 -0.2649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9467 -0.2649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0249 -1.0579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0249 -1.0579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7568 -1.7179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7568 -1.7179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1392 -2.3478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1392 -2.3478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9706 -1.7179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9706 -1.7179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5781 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5781 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7929 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7929 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4528 -1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4528 -1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2275 -1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2275 -1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5676 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5676 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2275 -2.9870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2275 -2.9870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4528 -2.9870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4528 -2.9870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2469 -2.3978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2469 -2.3978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4608 2.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4608 2.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4401 3.7246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4401 3.7246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 16 44 1 0 0 0 0 | + | 16 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 44 45 | + | M SAL 1 2 44 45 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 48 0.9229 2.987 | + | M SVB 1 48 0.9229 2.987 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGL0038 | + | ID FL5FADGL0038 |
| − | KNApSAcK_ID C00006009 | + | KNApSAcK_ID C00006009 |
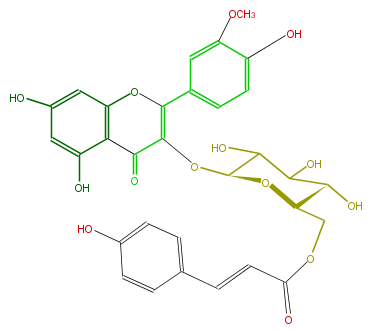
| − | NAME Isorhamnetin 3-(6''-p-coumarylglucoside) | + | NAME Isorhamnetin 3-(6''-p-coumarylglucoside) |
| − | CAS_RN 84534-24-7,110351-14-9 | + | CAS_RN 84534-24-7,110351-14-9 |
| − | FORMULA C31H28O14 | + | FORMULA C31H28O14 |
| − | EXACTMASS 624.147905604 | + | EXACTMASS 624.147905604 |
| − | AVERAGEMASS 624.5456200000001 | + | AVERAGEMASS 624.5456200000001 |
| − | SMILES [C@@H]([C@H]1COC(C=Cc(c5)ccc(O)c5)=O)(C(C([C@H](OC(=C3c(c4)cc(c(c4)O)OC)C(c(c(O)2)c(O3)cc(O)c2)=O)O1)O)O)O | + | SMILES [C@@H]([C@H]1COC(C=Cc(c5)ccc(O)c5)=O)(C(C([C@H](OC(=C3c(c4)cc(c(c4)O)OC)C(c(c(O)2)c(O3)cc(O)c2)=O)O1)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-2.9580 -0.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6866 -1.0953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0467 -1.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6782 -0.6251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9497 -0.0430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5896 0.0130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0383 -0.6811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6699 -0.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9413 0.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5813 0.4832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8266 -1.1349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5730 0.9532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0792 0.8962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4547 1.4325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1780 2.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4742 2.0829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8497 1.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1501 -0.3798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2099 0.4065 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.8223 -0.2649 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.5678 -0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3177 -0.2649 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7055 0.4065 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.9600 0.1935 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.4829 0.2117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2288 0.6591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3197 0.2419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7516 2.8450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7753 -1.7331 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5956 -0.5915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9467 -0.2649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0249 -1.0579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7568 -1.7179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1392 -2.3478 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9706 -1.7179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5781 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7929 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4528 -1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2275 -1.8087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5676 -2.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2275 -2.9870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4528 -2.9870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2469 -2.3978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4608 2.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4401 3.7246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 37 1 0 0 0 0
40 43 1 0 0 0 0
16 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 44 45
M SBL 1 1 48
M SMT 1 OCH3
M SVB 1 48 0.9229 2.987
S SKP 8
ID FL5FADGL0038
KNApSAcK_ID C00006009
NAME Isorhamnetin 3-(6''-p-coumarylglucoside)
CAS_RN 84534-24-7,110351-14-9
FORMULA C31H28O14
EXACTMASS 624.147905604
AVERAGEMASS 624.5456200000001
SMILES [C@@H]([C@H]1COC(C=Cc(c5)ccc(O)c5)=O)(C(C([C@H](OC(=C3c(c4)cc(c(c4)O)OC)C(c(c(O)2)c(O3)cc(O)c2)=O)O1)O)O)O
M END