Mol:FL5FAGGS0015
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3376 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3376 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3376 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3376 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7813 0.3325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7813 0.3325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2250 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2250 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2250 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2250 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7813 1.6172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7813 1.6172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6687 0.3325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6687 0.3325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1124 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1124 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1124 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1124 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6687 1.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6687 1.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6687 -0.1684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6687 -0.1684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4120 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4120 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1550 1.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1550 1.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7220 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7220 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7220 2.3955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7220 2.3955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1550 2.7228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1550 2.7228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4120 2.3955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4120 2.3955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7813 -0.3097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7813 -0.3097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3288 2.7458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3288 2.7458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8326 1.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8326 1.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5491 0.2441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5491 0.2441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1550 3.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1550 3.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5036 -0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5036 -0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1318 -1.1711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1318 -1.1711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9325 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9325 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1318 0.2281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1318 0.2281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5036 0.5909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5036 0.5909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7029 -0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7029 -0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4732 -0.1067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4732 -0.1067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6649 -1.4106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6649 -1.4106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9802 -1.7368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9802 -1.7368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5527 -0.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5527 -0.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3194 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3194 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0125 -2.8558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0125 -2.8558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0085 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0085 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3125 -2.8510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3125 -2.8510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9207 -2.8510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9207 -2.8510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2248 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2248 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9207 -1.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9207 -1.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3125 -1.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3125 -1.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2246 -1.2713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2246 -1.2713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8326 -2.3243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8326 -2.3243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2246 -3.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2246 -3.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2888 1.4136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2888 1.4136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 23 1 1 0 0 0 | + | 28 23 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 27 21 1 0 0 0 0 | + | 27 21 1 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | 14 44 1 0 0 0 0 | + | 14 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAGGS0015 | + | ID FL5FAGGS0015 |
| − | KNApSAcK_ID C00006044 | + | KNApSAcK_ID C00006044 |
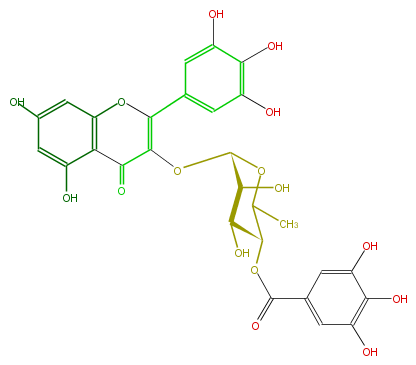
| − | NAME Myricetin 3-(4''-galloylrhamnoside) | + | NAME Myricetin 3-(4''-galloylrhamnoside) |
| − | CAS_RN 85541-03-3 | + | CAS_RN 85541-03-3 |
| − | FORMULA C28H24O16 | + | FORMULA C28H24O16 |
| − | EXACTMASS 616.1064347199999 | + | EXACTMASS 616.1064347199999 |
| − | AVERAGEMASS 616.48056 | + | AVERAGEMASS 616.48056 |
| − | SMILES O=C(OC(C(O)2)C(C)OC(OC(C5=O)=C(Oc(c54)cc(cc(O)4)O)c(c3)cc(c(O)c3O)O)C2O)c(c1)cc(O)c(c(O)1)O | + | SMILES O=C(OC(C(O)2)C(C)OC(OC(C5=O)=C(Oc(c54)cc(cc(O)4)O)c(c3)cc(c(O)c3O)O)C2O)c(c1)cc(O)c(c(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-3.3376 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3376 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7813 0.3325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2250 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2250 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7813 1.6172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6687 0.3325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1124 0.6536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1124 1.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6687 1.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6687 -0.1684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4120 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1550 1.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7220 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7220 2.3955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1550 2.7228 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4120 2.3955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7813 -0.3097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3288 2.7458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8326 1.6172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5491 0.2441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1550 3.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5036 -0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1318 -1.1711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9325 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1318 0.2281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5036 0.5909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7029 -0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4732 -0.1067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6649 -1.4106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9802 -1.7368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5527 -0.8317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3194 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0125 -2.8558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0085 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3125 -2.8510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9207 -2.8510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2248 -2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9207 -1.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3125 -1.7976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2246 -1.2713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8326 -2.3243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2246 -3.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2888 1.4136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
16 22 1 0 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 1 0 0 0
28 23 1 1 0 0 0
28 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
27 21 1 0 0 0 0
31 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 35 1 0 0 0 0
39 41 1 0 0 0 0
38 42 1 0 0 0 0
37 43 1 0 0 0 0
14 44 1 0 0 0 0
S SKP 8
ID FL5FAGGS0015
KNApSAcK_ID C00006044
NAME Myricetin 3-(4''-galloylrhamnoside)
CAS_RN 85541-03-3
FORMULA C28H24O16
EXACTMASS 616.1064347199999
AVERAGEMASS 616.48056
SMILES O=C(OC(C(O)2)C(C)OC(OC(C5=O)=C(Oc(c54)cc(cc(O)4)O)c(c3)cc(c(O)c3O)O)C2O)c(c1)cc(O)c(c(O)1)O
M END