Mol:FL5FECGA0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.8858 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8858 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8858 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8858 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3295 -1.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3295 -1.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7732 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7732 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7732 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7732 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3295 0.1904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3295 0.1904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2168 -1.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2168 -1.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6605 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6605 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6605 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6605 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2168 0.1904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2168 0.1904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2168 -1.5951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2168 -1.5951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1044 0.1903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1044 0.1903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4625 -0.1370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4625 -0.1370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0295 0.1903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0295 0.1903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0295 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0295 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4625 1.1723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4625 1.1723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1044 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1044 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4419 0.1903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4419 0.1903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0124 -1.3234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0124 -1.3234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0330 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0330 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6454 -1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6454 -1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3908 -1.2904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3908 -1.2904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1408 -1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1408 -1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5285 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5285 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7830 -1.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7830 -1.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3059 -1.0268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3059 -1.0268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0518 -0.5794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0518 -0.5794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5963 1.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5963 1.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3295 -1.7364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3295 -1.7364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4625 1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4625 1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6394 -0.4181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6394 -0.4181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6002 -0.6817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6002 -0.6817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8858 -1.0942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8858 -1.0942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8858 -1.4143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8858 -1.4143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6002 -1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6002 -1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 21 19 1 0 0 0 0 | + | 21 19 1 0 0 0 0 |
| − | 19 8 1 0 0 0 0 | + | 19 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 16 30 1 0 0 0 0 | + | 16 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 2 32 1 0 0 0 0 | + | 2 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 23 34 1 0 0 0 0 | + | 23 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 32 33 | + | M SAL 1 2 32 33 |
| − | M SBL 1 1 35 | + | M SBL 1 1 35 |
| − | M SMT 1 ^OCH3 | + | M SMT 1 ^OCH3 |
| − | M SBV 1 35 -6.0092 5.4263 | + | M SBV 1 35 -6.0092 5.4263 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 34 35 | + | M SAL 2 2 34 35 |
| − | M SBL 2 1 37 | + | M SBL 2 1 37 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 37 -4.5498 5.4240 | + | M SBV 2 37 -4.5498 5.4240 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FECGA0001 | + | ID FL5FECGA0001 |
| − | KNApSAcK_ID C00005638 | + | KNApSAcK_ID C00005638 |
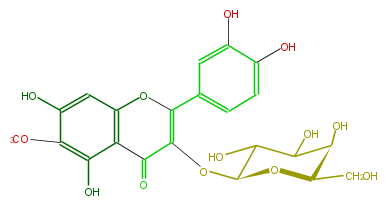
| − | NAME Patuletin 3-galactoside | + | NAME Patuletin 3-galactoside |
| − | CAS_RN 35399-34-9 | + | CAS_RN 35399-34-9 |
| − | FORMULA C22H22O13 | + | FORMULA C22H22O13 |
| − | EXACTMASS 494.10604078999995 | + | EXACTMASS 494.10604078999995 |
| − | AVERAGEMASS 494.40228 | + | AVERAGEMASS 494.40228 |
| − | SMILES O(C(=C(c(c4)cc(c(O)c4)O)3)C(=O)c(c2O3)c(c(OC)c(c2)O)O)C(O1)C(O)C(C(C(CO)1)O)O | + | SMILES O(C(=C(c(c4)cc(c(O)c4)O)3)C(=O)c(c2O3)c(c(OC)c(c2)O)O)C(O1)C(O)C(C(C(CO)1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-2.8858 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8858 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3295 -1.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7732 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7732 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3295 0.1904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2168 -1.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6605 -0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6605 -0.1308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2168 0.1904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2168 -1.5951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1044 0.1903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4625 -0.1370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0295 0.1903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0295 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4625 1.1723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1044 0.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4419 0.1903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0124 -1.3234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0330 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6454 -1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3908 -1.2904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1408 -1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5285 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7830 -1.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3059 -1.0268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0518 -0.5794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5963 1.1722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3295 -1.7364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4625 1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6394 -0.4181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6002 -0.6817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8858 -1.0942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8858 -1.4143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6002 -1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
25 27 1 0 0 0 0
21 19 1 0 0 0 0
19 8 1 0 0 0 0
23 22 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
16 30 1 0 0 0 0
24 31 1 0 0 0 0
2 32 1 0 0 0 0
32 33 1 0 0 0 0
23 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 32 33
M SBL 1 1 35
M SMT 1 ^OCH3
M SBV 1 35 -6.0092 5.4263
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 34 35
M SBL 2 1 37
M SMT 2 CH2OH
M SBV 2 37 -4.5498 5.4240
S SKP 8
ID FL5FECGA0001
KNApSAcK_ID C00005638
NAME Patuletin 3-galactoside
CAS_RN 35399-34-9
FORMULA C22H22O13
EXACTMASS 494.10604078999995
AVERAGEMASS 494.40228
SMILES O(C(=C(c(c4)cc(c(O)c4)O)3)C(=O)c(c2O3)c(c(OC)c(c2)O)O)C(O1)C(O)C(C(C(CO)1)O)O
M END