Mol:FL5FECGS0025
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 37 40 0 0 0 0 0 0 0 0999 V2000 | + | 37 40 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8182 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8182 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8182 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8182 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1037 -1.7392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1037 -1.7392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6107 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6107 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6107 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6107 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1037 -0.0892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1037 -0.0892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3252 -1.7392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3252 -1.7392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0397 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0397 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0397 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0397 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3252 -0.0892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3252 -0.0892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3252 -2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3252 -2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7539 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7539 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4821 -0.5098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4821 -0.5098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2103 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2103 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2103 0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2103 0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4821 1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4821 1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7539 0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7539 0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5324 -0.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5324 -0.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1037 -2.5638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1037 -2.5638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8153 1.1420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8153 1.1420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3277 0.0007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3277 0.0007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7793 -0.7230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7793 -0.7230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9897 -0.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9897 -0.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2279 -0.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2279 -0.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7815 0.1460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7815 0.1460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5881 -0.1436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5881 -0.1436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8153 -0.1829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8153 -0.1829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4472 -0.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4472 -0.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3078 -0.7657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3078 -0.7657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7828 -1.7015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7828 -1.7015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9019 -2.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9019 -2.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4762 1.8278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4762 1.8278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0431 2.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0431 2.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4249 -1.6644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4249 -1.6644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8728 -2.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8728 -2.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0538 0.3748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0538 0.3748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1937 1.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1937 1.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 8 30 1 0 0 0 0 | + | 8 30 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 16 32 1 0 0 0 0 | + | 16 32 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 2 34 1 0 0 0 0 | + | 2 34 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 26 36 1 0 0 0 0 | + | 26 36 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 30 31 | + | M SAL 1 2 30 31 |
| − | M SBL 1 1 34 | + | M SBL 1 1 34 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 34 -0.7432 0.3748 | + | M SBV 1 34 -0.7432 0.3748 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 32 33 | + | M SAL 2 2 32 33 |
| − | M SBL 2 1 36 | + | M SBL 2 1 36 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 36 0.0059 -0.6560 | + | M SBV 2 36 0.0059 -0.6560 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 34 35 | + | M SAL 3 2 34 35 |
| − | M SBL 3 1 38 | + | M SBL 3 1 38 |
| − | M SMT 3 ^ OCH3 | + | M SMT 3 ^ OCH3 |
| − | M SBV 3 38 0.6068 0.3377 | + | M SBV 3 38 0.6068 0.3377 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 36 37 | + | M SAL 4 2 36 37 |
| − | M SBL 4 1 40 | + | M SBL 4 1 40 |
| − | M SMT 4 ^ CH2OH | + | M SMT 4 ^ CH2OH |
| − | M SBV 4 40 0.4657 -0.5184 | + | M SBV 4 40 0.4657 -0.5184 |
| − | S SKP 5 | + | S SKP 5 |
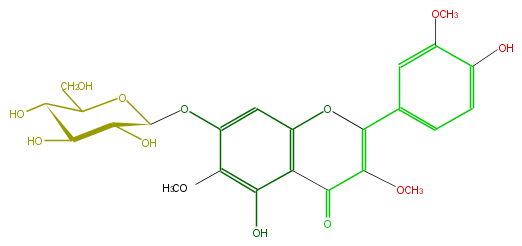
| − | ID FL5FECGS0025 | + | ID FL5FECGS0025 |
| − | FORMULA C24H26O13 | + | FORMULA C24H26O13 |
| − | EXACTMASS 522.137340918 | + | EXACTMASS 522.137340918 |
| − | AVERAGEMASS 522.45544 | + | AVERAGEMASS 522.45544 |
| − | SMILES c(c4O)(C2=O)c(cc(c4OC)OC(C3O)OC(CO)C(O)C3O)OC(=C(OC)2)c(c1)cc(c(c1)O)OC | + | SMILES c(c4O)(C2=O)c(cc(c4OC)OC(C3O)OC(CO)C(O)C3O)OC(=C(OC)2)c(c1)cc(c(c1)O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
37 40 0 0 0 0 0 0 0 0999 V2000
-0.8182 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8182 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1037 -1.7392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6107 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6107 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1037 -0.0892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3252 -1.7392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0397 -1.3267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0397 -0.5017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3252 -0.0892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3252 -2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7539 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4821 -0.5098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2103 -0.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2103 0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4821 1.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7539 0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5324 -0.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1037 -2.5638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8153 1.1420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3277 0.0007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7793 -0.7230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9897 -0.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2279 -0.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7815 0.1460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5881 -0.1436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8153 -0.1829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4472 -0.7247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3078 -0.7657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7828 -1.7015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9019 -2.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4762 1.8278 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0431 2.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4249 -1.6644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8728 -2.6245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0538 0.3748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1937 1.3285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
30 31 1 0 0 0 0
8 30 1 0 0 0 0
32 33 1 0 0 0 0
16 32 1 0 0 0 0
34 35 1 0 0 0 0
2 34 1 0 0 0 0
36 37 1 0 0 0 0
26 36 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 30 31
M SBL 1 1 34
M SMT 1 OCH3
M SBV 1 34 -0.7432 0.3748
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 32 33
M SBL 2 1 36
M SMT 2 OCH3
M SBV 2 36 0.0059 -0.6560
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 34 35
M SBL 3 1 38
M SMT 3 ^ OCH3
M SBV 3 38 0.6068 0.3377
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 36 37
M SBL 4 1 40
M SMT 4 ^ CH2OH
M SBV 4 40 0.4657 -0.5184
S SKP 5
ID FL5FECGS0025
FORMULA C24H26O13
EXACTMASS 522.137340918
AVERAGEMASS 522.45544
SMILES c(c4O)(C2=O)c(cc(c4OC)OC(C3O)OC(CO)C(O)C3O)OC(=C(OC)2)c(c1)cc(c(c1)O)OC
M END