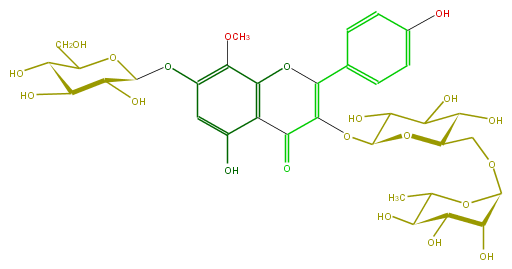
Mol:FL5FFAGL0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2644 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2644 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2644 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2644 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5534 -0.3239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5534 -0.3239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1577 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1577 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1577 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1577 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5534 1.3181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5534 1.3181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8687 -0.3239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8687 -0.3239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5797 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5797 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5797 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5797 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8687 1.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8687 1.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8687 -1.1115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8687 -1.1115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2905 1.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2905 1.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0150 0.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0150 0.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7397 1.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7397 1.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7397 2.1547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7397 2.1547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0150 2.5730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0150 2.5730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2905 2.1547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2905 2.1547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9751 1.3179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9751 1.3179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3000 -0.3483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3000 -0.3483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5534 -1.1445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5534 -1.1445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3089 -1.7308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3089 -1.7308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8798 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8798 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7050 -2.2382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7050 -2.2382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5349 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5349 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9641 -1.7308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9641 -1.7308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1390 -1.9666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1390 -1.9666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6177 -1.8006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6177 -1.8006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3193 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3193 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8903 -0.5569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8903 -0.5569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7154 -0.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7154 -0.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5454 -0.5569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5454 -0.5569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9745 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9745 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1494 -0.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1494 -0.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5857 0.0951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5857 0.0951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6761 0.5582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6761 0.5582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7528 0.0406 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7528 0.0406 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2923 -0.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2923 -0.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7477 -1.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7477 -1.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2637 -2.2376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2637 -2.2376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2914 -2.8606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2914 -2.8606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5349 -3.1980 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5349 -3.1980 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4873 2.5863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4873 2.5863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8208 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8208 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2752 0.6679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2752 0.6679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4895 0.9735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4895 0.9735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7313 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7313 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2823 1.5328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2823 1.5328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0849 1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0849 1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5211 1.1596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5211 1.1596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2427 0.6277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2427 0.6277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7698 0.5050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7698 0.5050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5534 2.0481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5534 2.0481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0046 3.1980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0046 3.1980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7142 1.8614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7142 1.8614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9641 2.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9641 2.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 29 19 1 0 0 0 0 | + | 29 19 1 0 0 0 0 |
| − | 42 15 1 0 0 0 0 | + | 42 15 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 18 1 0 0 0 0 | + | 46 18 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 6 52 1 0 0 0 0 | + | 6 52 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 48 54 1 0 0 0 0 | + | 48 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 58 0.0000 -0.7301 | + | M SBV 1 58 0.0000 -0.7301 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 54 55 | + | M SAL 2 2 54 55 |
| − | M SBL 2 1 60 | + | M SBL 2 1 60 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 60 0.6292 -0.6169 | + | M SBV 2 60 0.6292 -0.6169 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FFAGL0010 | + | ID FL5FFAGL0010 |
| − | FORMULA C34H42O21 | + | FORMULA C34H42O21 |
| − | EXACTMASS 786.2218584059999 | + | EXACTMASS 786.2218584059999 |
| − | AVERAGEMASS 786.68468 | + | AVERAGEMASS 786.68468 |
| − | SMILES c(c6)(c(c(O4)c(c(O)6)C(=O)C(=C4c(c5)ccc(c5)O)OC(O2)C(C(C(O)C(COC(C3O)OC(C)C(C3O)O)2)O)O)OC)OC(O1)C(O)C(C(C1CO)O)O | + | SMILES c(c6)(c(c(O4)c(c(O)6)C(=O)C(=C4c(c5)ccc(c5)O)OC(O2)C(C(C(O)C(COC(C3O)OC(C)C(C3O)O)2)O)O)OC)OC(O1)C(O)C(C(C1CO)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-1.2644 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2644 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5534 -0.3239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1577 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1577 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5534 1.3181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8687 -0.3239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5797 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5797 0.9076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8687 1.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8687 -1.1115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2905 1.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0150 0.8996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7397 1.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7397 2.1547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0150 2.5730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2905 2.1547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9751 1.3179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3000 -0.3483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5534 -1.1445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3089 -1.7308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8798 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7050 -2.2382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5349 -2.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9641 -1.7308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1390 -1.9666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6177 -1.8006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3193 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8903 -0.5569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7154 -0.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5454 -0.5569 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9745 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1494 -0.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5857 0.0951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6761 0.5582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7528 0.0406 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2923 -0.3933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7477 -1.0122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2637 -2.2376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2914 -2.8606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5349 -3.1980 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4873 2.5863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8208 1.3882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2752 0.6679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4895 0.9735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7313 0.9817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2823 1.5328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0849 1.2446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5211 1.1596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2427 0.6277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7698 0.5050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5534 2.0481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0046 3.1980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7142 1.8614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9641 2.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
33 35 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
25 38 1 0 0 0 0
37 38 1 0 0 0 0
22 39 1 0 0 0 0
23 40 1 0 0 0 0
24 41 1 0 0 0 0
29 19 1 0 0 0 0
42 15 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 18 1 0 0 0 0
52 53 1 0 0 0 0
6 52 1 0 0 0 0
54 55 1 0 0 0 0
48 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 58
M SMT 1 OCH3
M SBV 1 58 0.0000 -0.7301
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 54 55
M SBL 2 1 60
M SMT 2 ^ CH2OH
M SBV 2 60 0.6292 -0.6169
S SKP 5
ID FL5FFAGL0010
FORMULA C34H42O21
EXACTMASS 786.2218584059999
AVERAGEMASS 786.68468
SMILES c(c6)(c(c(O4)c(c(O)6)C(=O)C(=C4c(c5)ccc(c5)O)OC(O2)C(C(C(O)C(COC(C3O)OC(C)C(C3O)O)2)O)O)OC)OC(O1)C(O)C(C(C1CO)O)O
M END