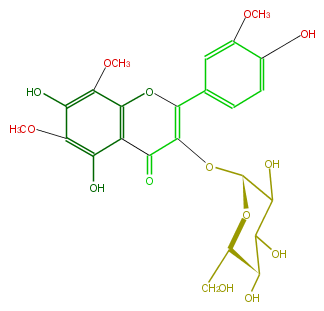
Mol:FL5FGCGL0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.1994 -5.1694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1994 -5.1694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8320 -5.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8320 -5.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0517 -4.4543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0517 -4.4543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6388 -3.9622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6388 -3.9622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0062 -4.0737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0062 -4.0737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2135 -4.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2135 -4.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8585 -3.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8585 -3.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4456 -2.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4456 -2.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1870 -2.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1870 -2.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4067 -3.5817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4067 -3.5817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3517 -3.2715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3517 -3.2715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5998 -2.4861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5998 -2.4861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3758 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3758 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7967 -1.3695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7967 -1.3695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4414 -1.4831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4414 -1.4831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6653 -2.0984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6653 -2.0984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2445 -2.5998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2445 -2.5998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0202 -5.7728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0202 -5.7728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8750 -2.1327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8750 -2.1327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3257 -0.7821 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.3257 -0.7821 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9381 -1.4536 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9381 -1.4536 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.6835 -1.2405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6835 -1.2405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4335 -1.4536 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4335 -1.4536 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8212 -0.7821 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8212 -0.7821 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0757 -0.9951 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.0757 -0.9951 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.5986 -0.9770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5986 -0.9770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3445 -0.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3445 -0.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3075 -0.9124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3075 -0.9124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0842 -0.7171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0842 -0.7171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6840 -4.3427 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6840 -4.3427 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8261 -4.5031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8261 -4.5031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7879 -4.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7879 -4.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2737 -1.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2737 -1.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4076 -2.7893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4076 -2.7893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2818 -1.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2818 -1.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2422 -1.6404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2422 -1.6404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7406 -5.7724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7406 -5.7724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2710 -6.4043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2710 -6.4043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 21 19 1 0 0 0 0 | + | 21 19 1 0 0 0 0 |
| − | 19 8 1 0 0 0 0 | + | 19 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 15 29 1 0 0 0 0 | + | 15 29 1 0 0 0 0 |
| − | 3 30 1 0 0 0 0 | + | 3 30 1 0 0 0 0 |
| − | 6 31 1 0 0 0 0 | + | 6 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 23 33 1 0 0 0 0 | + | 23 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 16 35 1 0 0 0 0 | + | 16 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 2 37 1 0 0 0 0 | + | 2 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 33 34 | + | M SAL 4 2 33 34 |
| − | M SBL 4 1 36 | + | M SBL 4 1 36 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 36 3.2737 -1.7983 | + | M SVB 4 36 3.2737 -1.7983 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 37 38 | + | M SAL 3 2 37 38 |
| − | M SBL 3 1 40 | + | M SBL 3 1 40 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 40 -3.3075 -0.6318 | + | M SVB 3 40 -3.3075 -0.6318 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 35 36 | + | M SAL 2 2 35 36 |
| − | M SBL 2 1 38 | + | M SBL 2 1 38 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 38 1.0386 1.7983 | + | M SVB 2 38 1.0386 1.7983 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 31 32 | + | M SAL 1 2 31 32 |
| − | M SBL 1 1 34 | + | M SBL 1 1 34 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 34 -1.7587 0.8133 | + | M SVB 1 34 -1.7587 0.8133 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FGCGL0001 | + | ID FL5FGCGL0001 |
| − | KNApSAcK_ID C00005783 | + | KNApSAcK_ID C00005783 |
| − | NAME Limocitrol 3-glucoside | + | NAME Limocitrol 3-glucoside |
| − | CAS_RN 77133-42-7 | + | CAS_RN 77133-42-7 |
| − | FORMULA C24H26O14 | + | FORMULA C24H26O14 |
| − | EXACTMASS 538.13225554 | + | EXACTMASS 538.13225554 |
| − | AVERAGEMASS 538.45484 | + | AVERAGEMASS 538.45484 |
| − | SMILES C(O4)(=C(C(c(c43)c(O)c(OC)c(O)c3OC)=O)O[C@H](O2)C(O)C([C@@H](O)[C@@H](CO)2)O)c(c1)cc(OC)c(c1)O | + | SMILES C(O4)(=C(C(c(c43)c(O)c(OC)c(O)c3OC)=O)O[C@H](O2)C(O)C([C@@H](O)[C@@H](CO)2)O)c(c1)cc(OC)c(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
0.1994 -5.1694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8320 -5.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0517 -4.4543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6388 -3.9622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0062 -4.0737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2135 -4.6773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8585 -3.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4456 -2.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1870 -2.9780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4067 -3.5817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3517 -3.2715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5998 -2.4861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3758 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7967 -1.3695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4414 -1.4831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6653 -2.0984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2445 -2.5998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0202 -5.7728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8750 -2.1327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3257 -0.7821 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9381 -1.4536 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6835 -1.2405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4335 -1.4536 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8212 -0.7821 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0757 -0.9951 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.5986 -0.9770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3445 -0.5295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3075 -0.9124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0842 -0.7171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6840 -4.3427 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8261 -4.5031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7879 -4.2294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2737 -1.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4076 -2.7893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2818 -1.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2422 -1.6404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7406 -5.7724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2710 -6.4043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
21 19 1 0 0 0 0
19 8 1 0 0 0 0
23 22 1 1 0 0 0
15 29 1 0 0 0 0
3 30 1 0 0 0 0
6 31 1 0 0 0 0
31 32 1 0 0 0 0
23 33 1 0 0 0 0
33 34 1 0 0 0 0
16 35 1 0 0 0 0
35 36 1 0 0 0 0
2 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 33 34
M SBL 4 1 36
M SMT 4 CH2OH
M SVB 4 36 3.2737 -1.7983
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 37 38
M SBL 3 1 40
M SMT 3 OCH3
M SVB 3 40 -3.3075 -0.6318
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 35 36
M SBL 2 1 38
M SMT 2 OCH3
M SVB 2 38 1.0386 1.7983
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 31 32
M SBL 1 1 34
M SMT 1 OCH3
M SVB 1 34 -1.7587 0.8133
S SKP 8
ID FL5FGCGL0001
KNApSAcK_ID C00005783
NAME Limocitrol 3-glucoside
CAS_RN 77133-42-7
FORMULA C24H26O14
EXACTMASS 538.13225554
AVERAGEMASS 538.45484
SMILES C(O4)(=C(C(c(c43)c(O)c(OC)c(O)c3OC)=O)O[C@H](O2)C(O)C([C@@H](O)[C@@H](CO)2)O)c(c1)cc(OC)c(c1)O
M END