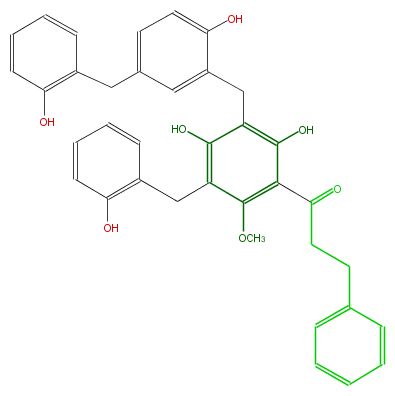
Mol:FL1DA9NC0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | 4.0476 -1.6296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0476 -1.6296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7142 -1.2448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7142 -1.2448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7142 -0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7142 -0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0476 -0.0903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0476 -0.0903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3811 -0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3811 -0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3811 -1.2448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3811 -1.2448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6729 -1.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6729 -1.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9235 -1.2210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9235 -1.2210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1995 -1.6390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1995 -1.6390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5125 -1.2424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5125 -1.2424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5125 -0.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5125 -0.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1667 -0.0660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1667 -0.0660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8459 -0.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8459 -0.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8459 -1.2424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8459 -1.2424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1667 -1.6345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1667 -1.6345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1995 -2.2407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1995 -2.2407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3781 -0.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3781 -0.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4016 -1.5632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4016 -1.5632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1667 -2.2237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1667 -2.2237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1207 -1.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1207 -1.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1207 -0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1207 -0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8046 0.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8046 0.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4885 -0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4885 -0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4885 -1.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4885 -1.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8046 -1.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8046 -1.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8046 -2.1085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8046 -2.1085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1667 0.6804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1667 0.6804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9148 1.1123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9148 1.1123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9148 1.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9148 1.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5560 2.2230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5560 2.2230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1973 1.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1973 1.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1973 1.1123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1973 1.1123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5560 0.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5560 0.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4269 1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4269 1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4269 1.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4269 1.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0705 2.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0705 2.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7142 1.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7142 1.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7142 1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7142 1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0705 0.7543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0705 0.7543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8922 2.1778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8922 2.1778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4148 2.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4148 2.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8046 0.6804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8046 0.6804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1111 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1111 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8256 -0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8256 -0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 9 16 2 0 0 0 0 | + | 9 16 2 0 0 0 0 |
| − | 13 17 1 0 0 0 0 | + | 13 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 18 20 1 0 0 0 0 | + | 18 20 1 0 0 0 0 |
| − | 20 21 2 0 0 0 0 | + | 20 21 2 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 12 27 1 0 0 0 0 | + | 12 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 2 0 0 0 0 | + | 28 29 2 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 29 41 1 0 0 0 0 | + | 29 41 1 0 0 0 0 |
| − | 22 42 1 0 0 0 0 | + | 22 42 1 0 0 0 0 |
| − | 42 34 1 0 0 0 0 | + | 42 34 1 0 0 0 0 |
| − | 11 43 1 0 0 0 0 | + | 11 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 -6.1860 4.3425 | + | M SBV 1 47 -6.1860 4.3425 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1DA9NC0010 | + | ID FL1DA9NC0010 |
| − | KNApSAcK_ID C00008120 | + | KNApSAcK_ID C00008120 |
| − | NAME Isotriuvaretin | + | NAME Isotriuvaretin |
| − | CAS_RN 137397-72-9 | + | CAS_RN 137397-72-9 |
| − | FORMULA C37H34O7 | + | FORMULA C37H34O7 |
| − | EXACTMASS 590.230453442 | + | EXACTMASS 590.230453442 |
| − | AVERAGEMASS 590.66166 | + | AVERAGEMASS 590.66166 |
| − | SMILES C(CCc(c5)cccc5)(c(c(OC)1)c(c(Cc(c(O)3)cc(Cc(c4)c(ccc4)O)cc3)c(c1Cc(c2)c(ccc2)O)O)O)=O | + | SMILES C(CCc(c5)cccc5)(c(c(OC)1)c(c(Cc(c(O)3)cc(Cc(c4)c(ccc4)O)cc3)c(c1Cc(c2)c(ccc2)O)O)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
4.0476 -1.6296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7142 -1.2448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7142 -0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0476 -0.0903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3811 -0.4751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3811 -1.2448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6729 -1.6537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9235 -1.2210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1995 -1.6390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5125 -1.2424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5125 -0.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1667 -0.0660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8459 -0.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8459 -1.2424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1667 -1.6345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1995 -2.2407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3781 -0.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4016 -1.5632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1667 -2.2237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1207 -1.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1207 -0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8046 0.0365 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4885 -0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4885 -1.1480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8046 -1.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8046 -2.1085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1667 0.6804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9148 1.1123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9148 1.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5560 2.2230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1973 1.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1973 1.1123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5560 0.7421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4269 1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4269 1.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0705 2.2407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7142 1.8691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7142 1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0705 0.7543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8922 2.1778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4148 2.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8046 0.6804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1111 -0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8256 -0.5250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
9 16 2 0 0 0 0
13 17 1 0 0 0 0
14 18 1 0 0 0 0
15 19 1 0 0 0 0
18 20 1 0 0 0 0
20 21 2 0 0 0 0
21 22 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 2 0 0 0 0
25 20 1 0 0 0 0
25 26 1 0 0 0 0
12 27 1 0 0 0 0
27 28 1 0 0 0 0
28 29 2 0 0 0 0
29 30 1 0 0 0 0
30 31 2 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
33 28 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
35 40 1 0 0 0 0
29 41 1 0 0 0 0
22 42 1 0 0 0 0
42 34 1 0 0 0 0
11 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 -6.1860 4.3425
S SKP 8
ID FL1DA9NC0010
KNApSAcK_ID C00008120
NAME Isotriuvaretin
CAS_RN 137397-72-9
FORMULA C37H34O7
EXACTMASS 590.230453442
AVERAGEMASS 590.66166
SMILES C(CCc(c5)cccc5)(c(c(OC)1)c(c(Cc(c(O)3)cc(Cc(c4)c(ccc4)O)cc3)c(c1Cc(c2)c(ccc2)O)O)O)=O
M END